BioImaging
-
 KAIST to Develop a Korean-style ChatGPT Platform Specifically Geared Toward Medical Diagnosis and Drug Discovery
On May 23rd, KAIST (President Kwang-Hyung Lee) announced that its Digital Bio-Health AI Research Center (Director: Professor JongChul Ye of KAIST Kim Jaechul Graduate School of AI) has been selected for the Ministry of Science and ICT's 'AI Top-Tier Young Researcher Support Program (AI Star Fellowship Project).' With a total investment of ₩11.5 billion from May 2025 to December 2030, the center will embark on the full-scale development of AI technology and a platform capable of independently inferring and determining the kinds of diseases, and discovering new drugs.
< Photo. On May 20th, a kick-off meeting for the AI Star Fellowship Project was held at KAIST Kim Jaechul Graduate School of AI’s Yangjae Research Center with the KAIST research team and participating organizations of Samsung Medical Center, NAVER Cloud, and HITS. [From left to right in the front row] Professor Jaegul Joo (KAIST), Professor Yoonjae Choi (KAIST), Professor Woo Youn Kim (KAIST/HITS), Professor JongChul Ye (KAIST), Professor Sungsoo Ahn (KAIST), Dr. Haanju Yoo (NAVER Cloud), Yoonho Lee (KAIST), HyeYoon Moon (Samsung Medical Center), Dr. Su Min Kim (Samsung Medical Center) >
This project aims to foster an innovative AI research ecosystem centered on young researchers and develop an inferential AI agent that can utilize and automatically expand specialized knowledge systems in the bio and medical fields.
Professor JongChul Ye of the Kim Jaechul Graduate School of AI will serve as the lead researcher, with young researchers from KAIST including Professors Yoonjae Choi, Kimin Lee, Sungsoo Ahn, and Chanyoung Park, along with mid-career researchers like Professors Jaegul Joo and Woo Youn Kim, jointly undertaking the project. They will collaborate with various laboratories within KAIST to conduct comprehensive research covering the entire cycle from the theoretical foundations of AI inference to its practical application.
Specifically, the main goals include: - Building high-performance inference models that integrate diverse medical knowledge systems to enhance the precision and reliability of diagnosis and treatment. - Developing a convergence inference platform that efficiently combines symbol-based inference with neural network models. - Securing AI technology for new drug development and biomarker discovery based on 'cell ontology.'
Furthermore, through close collaboration with industry and medical institutions such as Samsung Medical Center, NAVER Cloud, and HITS Co., Ltd., the project aims to achieve: - Clinical diagnostic AI utilizing medical knowledge systems. - AI-based molecular target exploration for new drug development. - Commercialization of an extendible AI inference platform.
Professor JongChul Ye, Director of KAIST's Digital Bio-Health AI Research Center, stated, "At a time when competition in AI inference model development is intensifying, it is a great honor for KAIST to lead the development of AI technology specialized in the bio and medical fields with world-class young researchers." He added, "We will do our best to ensure that the participating young researchers reach a world-leading level in terms of research achievements after the completion of this seven-year project starting in 2025."
The AI Star Fellowship is a newly established program where post-doctoral researchers and faculty members within seven years of appointment participate as project leaders (PLs) to independently lead research. Multiple laboratories within a university and demand-side companies form a consortium to operate the program.
Through this initiative, KAIST plans to nurture bio-medical convergence AI talent and simultaneously promote the commercialization of core technologies in collaboration with Samsung Medical Center, NAVER Cloud, and HITS.
2025.05.26 View 928
KAIST to Develop a Korean-style ChatGPT Platform Specifically Geared Toward Medical Diagnosis and Drug Discovery
On May 23rd, KAIST (President Kwang-Hyung Lee) announced that its Digital Bio-Health AI Research Center (Director: Professor JongChul Ye of KAIST Kim Jaechul Graduate School of AI) has been selected for the Ministry of Science and ICT's 'AI Top-Tier Young Researcher Support Program (AI Star Fellowship Project).' With a total investment of ₩11.5 billion from May 2025 to December 2030, the center will embark on the full-scale development of AI technology and a platform capable of independently inferring and determining the kinds of diseases, and discovering new drugs.
< Photo. On May 20th, a kick-off meeting for the AI Star Fellowship Project was held at KAIST Kim Jaechul Graduate School of AI’s Yangjae Research Center with the KAIST research team and participating organizations of Samsung Medical Center, NAVER Cloud, and HITS. [From left to right in the front row] Professor Jaegul Joo (KAIST), Professor Yoonjae Choi (KAIST), Professor Woo Youn Kim (KAIST/HITS), Professor JongChul Ye (KAIST), Professor Sungsoo Ahn (KAIST), Dr. Haanju Yoo (NAVER Cloud), Yoonho Lee (KAIST), HyeYoon Moon (Samsung Medical Center), Dr. Su Min Kim (Samsung Medical Center) >
This project aims to foster an innovative AI research ecosystem centered on young researchers and develop an inferential AI agent that can utilize and automatically expand specialized knowledge systems in the bio and medical fields.
Professor JongChul Ye of the Kim Jaechul Graduate School of AI will serve as the lead researcher, with young researchers from KAIST including Professors Yoonjae Choi, Kimin Lee, Sungsoo Ahn, and Chanyoung Park, along with mid-career researchers like Professors Jaegul Joo and Woo Youn Kim, jointly undertaking the project. They will collaborate with various laboratories within KAIST to conduct comprehensive research covering the entire cycle from the theoretical foundations of AI inference to its practical application.
Specifically, the main goals include: - Building high-performance inference models that integrate diverse medical knowledge systems to enhance the precision and reliability of diagnosis and treatment. - Developing a convergence inference platform that efficiently combines symbol-based inference with neural network models. - Securing AI technology for new drug development and biomarker discovery based on 'cell ontology.'
Furthermore, through close collaboration with industry and medical institutions such as Samsung Medical Center, NAVER Cloud, and HITS Co., Ltd., the project aims to achieve: - Clinical diagnostic AI utilizing medical knowledge systems. - AI-based molecular target exploration for new drug development. - Commercialization of an extendible AI inference platform.
Professor JongChul Ye, Director of KAIST's Digital Bio-Health AI Research Center, stated, "At a time when competition in AI inference model development is intensifying, it is a great honor for KAIST to lead the development of AI technology specialized in the bio and medical fields with world-class young researchers." He added, "We will do our best to ensure that the participating young researchers reach a world-leading level in terms of research achievements after the completion of this seven-year project starting in 2025."
The AI Star Fellowship is a newly established program where post-doctoral researchers and faculty members within seven years of appointment participate as project leaders (PLs) to independently lead research. Multiple laboratories within a university and demand-side companies form a consortium to operate the program.
Through this initiative, KAIST plans to nurture bio-medical convergence AI talent and simultaneously promote the commercialization of core technologies in collaboration with Samsung Medical Center, NAVER Cloud, and HITS.
2025.05.26 View 928 -
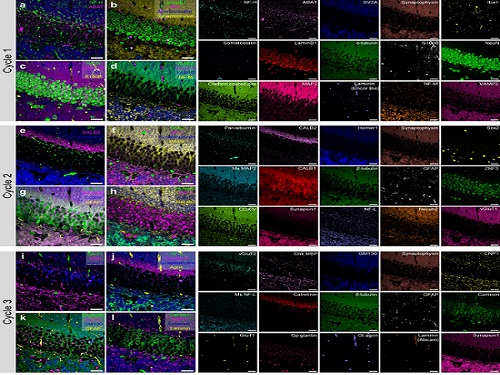 PICASSO Technique Drives Biological Molecules into Technicolor
The new imaging approach brings current imaging colors from four to more than 15 for mapping overlapping proteins
Pablo Picasso’s surreal cubist artistic style shifted common features into unrecognizable scenes, but a new imaging approach bearing his namesake may elucidate the most complicated subject: the brain. Employing artificial intelligence to clarify spectral color blending of tiny molecules used to stain specific proteins and other items of research interest, the PICASSO technique, allows researchers to use more than 15 colors to image and parse our overlapping proteins.
The PICASSO developers, based in Korea, published their approach on May 5 in Nature Communications.
Fluorophores — the staining molecules — emit specific colors when excited by a light, but if more than four fluorophores are used, their emitted colors overlap and blend. Researchers previously developed techniques to correct this spectral overlap by precisely defining the matrix of mixed and unmixed images. This measurement depends on reference spectra, found by identifying clear images of only one fluorophore-stained specimen or of multiple, identically prepared specimens that only contain a single fluorophore each.
“Such reference spectra measurement could be complicated to perform in highly heterogeneous specimens, such as the brain, due to the highly varied emission spectra of fluorophores depending on the subregions from which the spectra were measured,” said co-corresponding author Young-Gyu Yoon, professor in the School of Electrical Engineering at KAIST. He explained that the subregions would each need their own spectra reference measurements, making for an inefficient, time-consuming process. “To address this problem, we developed an approach that does not require reference spectra measurements.”
The approach is the “Process of ultra-multiplexed Imaging of biomolecules viA the unmixing of the Signals of Spectrally Overlapping fluorophores,” also known as PICASSO. Ultra-multiplexed imaging refers to visualizing the numerous individual components of a unit. Like a cinema multiplex in which each theater plays a different movie, each protein in a cell has a different role. By staining with fluorophores, researchers can begin to understand those roles.
“We devised a strategy based on information theory; unmixing is performed by iteratively minimizing the mutual information between mixed images,” said co-corresponding author Jae-Byum Chang, professor in the Department of Materials Science and Engineering, KAIST. “This allows us to get away with the assumption that the spatial distribution of different proteins is mutually exclusive and enables accurate information unmixing.”
To demonstrate PICASSO’s capabilities, the researchers applied the technique to imaging a mouse brain. With a single round of staining, they performed 15-color multiplexed imaging of a mouse brain. Although small, mouse brains are still complex, multifaceted organs that can take significant resources to map. According to the researchers, PICASSO can improve the capabilities of other imaging techniques and allow for the use of even more fluorophore colors.
Using one such imaging technique in combination with PICASSO, the team achieved 45-color multiplexed imaging of the mouse brain in only three staining and imaging cycles, according to Yoon.
“PICASSO is a versatile tool for the multiplexed biomolecule imaging of cultured cells, tissue slices and clinical specimens,” Chang said. “We anticipate that PICASSO will be useful for a broad range of applications for which biomolecules’ spatial information is important. One such application the tool would be useful for is revealing the cellular heterogeneities of tumor microenvironments, especially the heterogeneous populations of immune cells, which are closely related to cancer prognoses and the efficacy of cancer therapies.”
The Samsung Research Funding & Incubation Center for Future Technology supported this work. Spectral imaging was performed at the Korea Basic Science Institute Western Seoul Center.
-PublicationJunyoung Seo, Yeonbo Sim, Jeewon Kim, Hyunwoo Kim, In Cho, Hoyeon Nam, Yong-Gyu Yoon, Jae-Byum Chang, “PICASSO allows ultra-multiplexed fluorescence imaging of spatiallyoverlapping proteins without reference spectra measurements,” May 5, Nature Communications (doi.org/10.1038/s41467-022-30168-z)
-ProfileProfessor Jae-Byum ChangDepartment of Materials Science and EngineeringCollege of EngineeringKAIST
Professor Young-Gyu YoonSchool of Electrical EngineeringCollege of EngineeringKAIST
2022.06.22 View 10353
PICASSO Technique Drives Biological Molecules into Technicolor
The new imaging approach brings current imaging colors from four to more than 15 for mapping overlapping proteins
Pablo Picasso’s surreal cubist artistic style shifted common features into unrecognizable scenes, but a new imaging approach bearing his namesake may elucidate the most complicated subject: the brain. Employing artificial intelligence to clarify spectral color blending of tiny molecules used to stain specific proteins and other items of research interest, the PICASSO technique, allows researchers to use more than 15 colors to image and parse our overlapping proteins.
The PICASSO developers, based in Korea, published their approach on May 5 in Nature Communications.
Fluorophores — the staining molecules — emit specific colors when excited by a light, but if more than four fluorophores are used, their emitted colors overlap and blend. Researchers previously developed techniques to correct this spectral overlap by precisely defining the matrix of mixed and unmixed images. This measurement depends on reference spectra, found by identifying clear images of only one fluorophore-stained specimen or of multiple, identically prepared specimens that only contain a single fluorophore each.
“Such reference spectra measurement could be complicated to perform in highly heterogeneous specimens, such as the brain, due to the highly varied emission spectra of fluorophores depending on the subregions from which the spectra were measured,” said co-corresponding author Young-Gyu Yoon, professor in the School of Electrical Engineering at KAIST. He explained that the subregions would each need their own spectra reference measurements, making for an inefficient, time-consuming process. “To address this problem, we developed an approach that does not require reference spectra measurements.”
The approach is the “Process of ultra-multiplexed Imaging of biomolecules viA the unmixing of the Signals of Spectrally Overlapping fluorophores,” also known as PICASSO. Ultra-multiplexed imaging refers to visualizing the numerous individual components of a unit. Like a cinema multiplex in which each theater plays a different movie, each protein in a cell has a different role. By staining with fluorophores, researchers can begin to understand those roles.
“We devised a strategy based on information theory; unmixing is performed by iteratively minimizing the mutual information between mixed images,” said co-corresponding author Jae-Byum Chang, professor in the Department of Materials Science and Engineering, KAIST. “This allows us to get away with the assumption that the spatial distribution of different proteins is mutually exclusive and enables accurate information unmixing.”
To demonstrate PICASSO’s capabilities, the researchers applied the technique to imaging a mouse brain. With a single round of staining, they performed 15-color multiplexed imaging of a mouse brain. Although small, mouse brains are still complex, multifaceted organs that can take significant resources to map. According to the researchers, PICASSO can improve the capabilities of other imaging techniques and allow for the use of even more fluorophore colors.
Using one such imaging technique in combination with PICASSO, the team achieved 45-color multiplexed imaging of the mouse brain in only three staining and imaging cycles, according to Yoon.
“PICASSO is a versatile tool for the multiplexed biomolecule imaging of cultured cells, tissue slices and clinical specimens,” Chang said. “We anticipate that PICASSO will be useful for a broad range of applications for which biomolecules’ spatial information is important. One such application the tool would be useful for is revealing the cellular heterogeneities of tumor microenvironments, especially the heterogeneous populations of immune cells, which are closely related to cancer prognoses and the efficacy of cancer therapies.”
The Samsung Research Funding & Incubation Center for Future Technology supported this work. Spectral imaging was performed at the Korea Basic Science Institute Western Seoul Center.
-PublicationJunyoung Seo, Yeonbo Sim, Jeewon Kim, Hyunwoo Kim, In Cho, Hoyeon Nam, Yong-Gyu Yoon, Jae-Byum Chang, “PICASSO allows ultra-multiplexed fluorescence imaging of spatiallyoverlapping proteins without reference spectra measurements,” May 5, Nature Communications (doi.org/10.1038/s41467-022-30168-z)
-ProfileProfessor Jae-Byum ChangDepartment of Materials Science and EngineeringCollege of EngineeringKAIST
Professor Young-Gyu YoonSchool of Electrical EngineeringCollege of EngineeringKAIST
2022.06.22 View 10353