structural
-
 KAIST perfectly reproduces Joseon-era Irworobongdo without pigments
Typically, chemical pigments that absorb specific wavelengths of light within the visible spectrum are required to produce colors. However, KAIST researchers have successfully reproduced the Joseon-era Irworobongdo [일월오봉도] painting using ultra-precise color graphics without any chemical pigments, allowing for the permanent and eco-friendly preservation of color graphics without fading or discoloration.
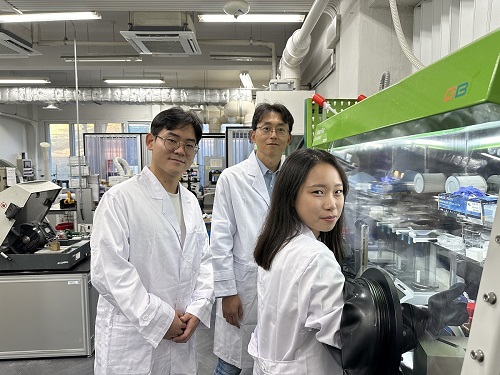
< (From left) Chaerim Son, a graduate of the Department of Biochemical Engineering (lead author), Seong Kyeong Nam, a graduate of the PhD program, Jiwoo Lee, a PhD student, and Professor Shin-Hyun Kim >
KAIST (represented by President Kwang Hyung Lee) announced on the 26th of February that a research team led by Professor Shinhyun Kim from the Department of Biological and Chemical Engineering had developed a technology that enables high-resolution color graphics without using any chemical pigments by employing hemisphere-shaped microstructures.
Morpho butterflies that are brilliant blue in color or Panther chameleons that change skin color exhibit coloration without chemical pigments, as ordered nanostructures within a material reflect visible light through optical interference. Since structural colors arise from physical structures rather than chemical substances, a single material can produce a wide range of colors.
However, the artificial implementation of structural coloration is highly challenging due to the complexity of creating ordered nanostructures. Additionally, it is difficult to produce a variety of colors and to pattern them precisely into complex designs.
< Figure 1. Principle of structural color expression using micro-hemispheres (left) and method of forming micro-hemisphere patterns based on photolithography (right) >
Professor Kim’s team overcame these challenges by using smooth-surfaced hemispherical microstructures instead of ordered nanostructures, enabling the high-precision patterning of diverse structural colors.
When light enters the inverted hemispherical microstructures, the portion of light entering from the sides undergoes total internal reflection along the curved surface, creating retroreflection. When the hemisphere diameter is approximately 10 micrometers (about one-tenth the thickness of a human hair), light traveling along different reflection paths interferes within the visible spectrum, producing structural coloration.
< Figure 2. “Irworobongdo”, the Painting of the Sun, Moon, and the Five Peaks, reproduced in fingernail size without pigment using approximately 200,000 micro-hemispheres >
The structural color can be tuned by adjusting the size of the hemispheres. By arranging hemispheres of varying sizes, much like mixing paints on a palette, an infinite range of colors can be generated.
To precisely pattern microscale hemispheres of different sizes, the research team employed photolithography* using positive photoresists** commonly used in semiconductor processing. They first patterned photoresists into micropillar structures, then induced reflow*** by heating the material, forming hemispherical microstructures.
*Photolithography: A technique used in semiconductor fabrication to pattern microscale structures.
**Positive photoresist: A photosensitive polymer that dissolves more easily in a developer solution after exposure to ultraviolet light.
***Reflow: A process in which a polymer material softens and reshapes into a curved structure when heated.
This method enables the formation of hemisphere-shaped microstructures with the desired sizes and colors in a single-step fabrication process. It also allows for the reproduction of arbitrary color graphics using a single material without any pigments.
The ultra-precise color graphics created with this technique can exhibit color variations depending on the angle of incident light or the viewing perspective. The pattern appears colored from one direction while remaining transparent from the opposite side, exhibiting a Janus effect. These structural color graphics achieve resolution comparable to cutting-edge LED displays, allowing complex color images to be captured within a fingernail-sized area and projected onto large screens.
< Figure 3. “Irworobongdo” that displays different shades depending on the angle of light and viewing direction >
Professor Shinhyun Kim, who led the research, stated, “Our newly developed pigment-free color graphics technology can serve as an innovative method for artistic expression, merging art with advanced materials. Additionally, it holds broad application potential in optical devices and sensors, anti-counterfeiting materials, aesthetic photocard printing, and many other fields.”
This research, with KAIST researcher Chaerim Son as the first author, was published in the prestigious materials science journal Advanced Materials on February 5.
(Paper title: “Retroreflective Multichrome Microdome Arrays Created by Single-Step Reflow”, DOI: 10.1002/adma.202413143 )
< Figure 4. Famous paintings reproduced without pigment: “Impression, Sunrise” (left), “Girl with a Pearl Earring” (right) >
The study was supported by the National Research Foundation of Korea through the Pioneer Converging Technology R&D Program and the Mid-Career Researcher Program.
2025.02.26 View 4162
KAIST perfectly reproduces Joseon-era Irworobongdo without pigments
Typically, chemical pigments that absorb specific wavelengths of light within the visible spectrum are required to produce colors. However, KAIST researchers have successfully reproduced the Joseon-era Irworobongdo [일월오봉도] painting using ultra-precise color graphics without any chemical pigments, allowing for the permanent and eco-friendly preservation of color graphics without fading or discoloration.
< (From left) Chaerim Son, a graduate of the Department of Biochemical Engineering (lead author), Seong Kyeong Nam, a graduate of the PhD program, Jiwoo Lee, a PhD student, and Professor Shin-Hyun Kim >
KAIST (represented by President Kwang Hyung Lee) announced on the 26th of February that a research team led by Professor Shinhyun Kim from the Department of Biological and Chemical Engineering had developed a technology that enables high-resolution color graphics without using any chemical pigments by employing hemisphere-shaped microstructures.
Morpho butterflies that are brilliant blue in color or Panther chameleons that change skin color exhibit coloration without chemical pigments, as ordered nanostructures within a material reflect visible light through optical interference. Since structural colors arise from physical structures rather than chemical substances, a single material can produce a wide range of colors.
However, the artificial implementation of structural coloration is highly challenging due to the complexity of creating ordered nanostructures. Additionally, it is difficult to produce a variety of colors and to pattern them precisely into complex designs.
< Figure 1. Principle of structural color expression using micro-hemispheres (left) and method of forming micro-hemisphere patterns based on photolithography (right) >
Professor Kim’s team overcame these challenges by using smooth-surfaced hemispherical microstructures instead of ordered nanostructures, enabling the high-precision patterning of diverse structural colors.
When light enters the inverted hemispherical microstructures, the portion of light entering from the sides undergoes total internal reflection along the curved surface, creating retroreflection. When the hemisphere diameter is approximately 10 micrometers (about one-tenth the thickness of a human hair), light traveling along different reflection paths interferes within the visible spectrum, producing structural coloration.
< Figure 2. “Irworobongdo”, the Painting of the Sun, Moon, and the Five Peaks, reproduced in fingernail size without pigment using approximately 200,000 micro-hemispheres >
The structural color can be tuned by adjusting the size of the hemispheres. By arranging hemispheres of varying sizes, much like mixing paints on a palette, an infinite range of colors can be generated.
To precisely pattern microscale hemispheres of different sizes, the research team employed photolithography* using positive photoresists** commonly used in semiconductor processing. They first patterned photoresists into micropillar structures, then induced reflow*** by heating the material, forming hemispherical microstructures.
*Photolithography: A technique used in semiconductor fabrication to pattern microscale structures.
**Positive photoresist: A photosensitive polymer that dissolves more easily in a developer solution after exposure to ultraviolet light.
***Reflow: A process in which a polymer material softens and reshapes into a curved structure when heated.
This method enables the formation of hemisphere-shaped microstructures with the desired sizes and colors in a single-step fabrication process. It also allows for the reproduction of arbitrary color graphics using a single material without any pigments.
The ultra-precise color graphics created with this technique can exhibit color variations depending on the angle of incident light or the viewing perspective. The pattern appears colored from one direction while remaining transparent from the opposite side, exhibiting a Janus effect. These structural color graphics achieve resolution comparable to cutting-edge LED displays, allowing complex color images to be captured within a fingernail-sized area and projected onto large screens.
< Figure 3. “Irworobongdo” that displays different shades depending on the angle of light and viewing direction >
Professor Shinhyun Kim, who led the research, stated, “Our newly developed pigment-free color graphics technology can serve as an innovative method for artistic expression, merging art with advanced materials. Additionally, it holds broad application potential in optical devices and sensors, anti-counterfeiting materials, aesthetic photocard printing, and many other fields.”
This research, with KAIST researcher Chaerim Son as the first author, was published in the prestigious materials science journal Advanced Materials on February 5.
(Paper title: “Retroreflective Multichrome Microdome Arrays Created by Single-Step Reflow”, DOI: 10.1002/adma.202413143 )
< Figure 4. Famous paintings reproduced without pigment: “Impression, Sunrise” (left), “Girl with a Pearl Earring” (right) >
The study was supported by the National Research Foundation of Korea through the Pioneer Converging Technology R&D Program and the Mid-Career Researcher Program.
2025.02.26 View 4162 -
 KAIST Develops a Multifunctional Structural Battery Capable of Energy Storage and Load Support
Structural batteries are used in industries such as eco-friendly, energy-based automobiles, mobility, and aerospace, and they must simultaneously meet the requirements of high energy density for energy storage and high load-bearing capacity. Conventional structural battery technology has struggled to enhance both functions concurrently. However, KAIST researchers have succeeded in developing foundational technology to address this issue.
< Photo 1. (From left) Professor Seong Su Kim, PhD candidates Sangyoon Bae and Su Hyun Lim of the Department of Mechanical Engineering >
< Photo 2. (From left) Professor Seong Su Kim and Master's Graduate Mohamad A. Raja of KAIST Department of Mechanical Engineering >
KAIST (represented by President Kwang Hyung Lee) announced on the 19th of November that Professor Seong Su Kim's team from the Department of Mechanical Engineering has developed a thin, uniform, high-density, multifunctional structural carbon fiber composite battery* capable of supporting loads, and that is free from fire risks while offering high energy density.
*Multifunctional structural batteries: Refers to the ability of each material in the composite to simultaneously serve as a load-bearing structure and an energy storage element.
Early structural batteries involved embedding commercial lithium-ion batteries into layered composite materials. These batteries suffered from low integration of their mechanical and electrochemical properties, leading to challenges in material processing, assembly, and design optimization, making commercialization difficult.
To overcome these challenges, Professor Kim's team explored the concept of "energy-storing composite materials," focusing on interface and curing properties, which are critical in traditional composite design. This led to the development of high-density multifunctional structural carbon fiber composite batteries that maximize multifunctionality.
The team analyzed the curing mechanisms of epoxy resin, known for its strong mechanical properties, combined with ionic liquid and carbonate electrolyte-based solid polymer electrolytes. By controlling temperature and pressure, they were able to optimize the curing process.
The newly developed structural battery was manufactured through vacuum compression molding, increasing the volume fraction of carbon fibers—serving as both electrodes and current collectors—by over 160% compared to previous carbon-fiber-based batteries.
This greatly increased the contact area between electrodes and electrolytes, resulting in a high-density structural battery with improved electrochemical performance. Furthermore, the team effectively controlled air bubbles within the structural battery during the curing process, simultaneously enhancing the battery's mechanical properties.
Professor Seong Su Kim, the lead researcher, explained, “We proposed a framework for designing solid polymer electrolytes, a core material for high-stiffness, ultra-thin structural batteries, from both material and structural perspectives. These material-based structural batteries can serve as internal components in cars, drones, airplanes, and robots, significantly extending their operating time with a single charge. This represents a foundational technology for next-generation multifunctional energy storage applications.”
< Figure 2. Supplementary cover of ACS Applied Materials & Interfaces >
Mohamad A. Raja, a master’s graduate of KAIST’s Department of Mechanical Engineering, participated as the first author of this research, which was published in the prestigious journal ACS Applied Materials & Interfaces on September 10. The paper was recognized for its excellence and selected as a supplementary cover article. (Paper title: “Thin, Uniform, and Highly Packed Multifunctional Structural Carbon Fiber Composite Battery Lamina Informed by Solid Polymer Electrolyte Cure Kinetics.” https://doi.org/10.1021/acsami.4c08698)
This research was supported by the National Research Foundation of Korea’s Mid-Career Researcher Program and the National Semiconductor Research Laboratory Development Program.
2024.11.27 View 5781
KAIST Develops a Multifunctional Structural Battery Capable of Energy Storage and Load Support
Structural batteries are used in industries such as eco-friendly, energy-based automobiles, mobility, and aerospace, and they must simultaneously meet the requirements of high energy density for energy storage and high load-bearing capacity. Conventional structural battery technology has struggled to enhance both functions concurrently. However, KAIST researchers have succeeded in developing foundational technology to address this issue.
< Photo 1. (From left) Professor Seong Su Kim, PhD candidates Sangyoon Bae and Su Hyun Lim of the Department of Mechanical Engineering >
< Photo 2. (From left) Professor Seong Su Kim and Master's Graduate Mohamad A. Raja of KAIST Department of Mechanical Engineering >
KAIST (represented by President Kwang Hyung Lee) announced on the 19th of November that Professor Seong Su Kim's team from the Department of Mechanical Engineering has developed a thin, uniform, high-density, multifunctional structural carbon fiber composite battery* capable of supporting loads, and that is free from fire risks while offering high energy density.
*Multifunctional structural batteries: Refers to the ability of each material in the composite to simultaneously serve as a load-bearing structure and an energy storage element.
Early structural batteries involved embedding commercial lithium-ion batteries into layered composite materials. These batteries suffered from low integration of their mechanical and electrochemical properties, leading to challenges in material processing, assembly, and design optimization, making commercialization difficult.
To overcome these challenges, Professor Kim's team explored the concept of "energy-storing composite materials," focusing on interface and curing properties, which are critical in traditional composite design. This led to the development of high-density multifunctional structural carbon fiber composite batteries that maximize multifunctionality.
The team analyzed the curing mechanisms of epoxy resin, known for its strong mechanical properties, combined with ionic liquid and carbonate electrolyte-based solid polymer electrolytes. By controlling temperature and pressure, they were able to optimize the curing process.
The newly developed structural battery was manufactured through vacuum compression molding, increasing the volume fraction of carbon fibers—serving as both electrodes and current collectors—by over 160% compared to previous carbon-fiber-based batteries.
This greatly increased the contact area between electrodes and electrolytes, resulting in a high-density structural battery with improved electrochemical performance. Furthermore, the team effectively controlled air bubbles within the structural battery during the curing process, simultaneously enhancing the battery's mechanical properties.
Professor Seong Su Kim, the lead researcher, explained, “We proposed a framework for designing solid polymer electrolytes, a core material for high-stiffness, ultra-thin structural batteries, from both material and structural perspectives. These material-based structural batteries can serve as internal components in cars, drones, airplanes, and robots, significantly extending their operating time with a single charge. This represents a foundational technology for next-generation multifunctional energy storage applications.”
< Figure 2. Supplementary cover of ACS Applied Materials & Interfaces >
Mohamad A. Raja, a master’s graduate of KAIST’s Department of Mechanical Engineering, participated as the first author of this research, which was published in the prestigious journal ACS Applied Materials & Interfaces on September 10. The paper was recognized for its excellence and selected as a supplementary cover article. (Paper title: “Thin, Uniform, and Highly Packed Multifunctional Structural Carbon Fiber Composite Battery Lamina Informed by Solid Polymer Electrolyte Cure Kinetics.” https://doi.org/10.1021/acsami.4c08698)
This research was supported by the National Research Foundation of Korea’s Mid-Career Researcher Program and the National Semiconductor Research Laboratory Development Program.
2024.11.27 View 5781 -
 'Jumping Genes' Found to Alter Human Colon Genomes, Offering Insights into Aging and Tumorigenesis
The Korea Advanced Institute of Science and Technology (KAIST) and their collaborators have conducted a groundbreaking study targeting 'jumping genes' in the entire genomes of the human large intestine. Published in Nature on May 18 2023, the research unveils the surprising activity of 'Long interspersed nuclear element-1 (L1),' a type of jumping gene previously thought to be mostly dormant in human genomes. The study shows that L1 genes can become activated and disrupt genomic functions throughout an individual's lifetime, particularly in the colorectal epithelium.
(Paper Title: Widespread somatic L1 retrotransposition in normal colorectal epithelium, https://www.nature.com/articles/s41586-023-06046-z)
With approximately 500,000 L1 jumping genes, accounting for 17% of the human genome, they have long been recognized for their contribution to the evolution of the human species by introducing 'disruptive innovation' to genome sequences. Until now, it was believed that most L1 elements had lost their ability to jump in normal tissues of modern humans. However, this study reveals that some L1 jumping genes can be widely activated in normal cells, leading to the accumulation of genomic mutations over an individual's lifetime. The rate of L1 jumping and resulting genomic changes vary among different cell types, with a notable concentration observed in aged colon epithelial cells. The study illustrates that every colonic epithelial cell experiences an L1 jumping event by the age of 40 on average.
The research, led by co-first authors Chang Hyun Nam (a graduate student at KAIST) and Dr. Jeonghwan Youk (former graduate student at KAIST and assistant clinical professor at Seoul National University Hospital), involved the analysis of whole-genome sequences from 899 single cells obtained from skin (fibroblasts), blood, and colon epithelial tissues collected from 28 individuals. The study uncovers the activation of L1 jumping genes in normal cells, resulting in the gradual accumulation of genomic mutations over time. Additionally, the team explored epigenomic (DNA methylation) sequences to understand the mechanism behind L1 jumping gene activation. They found that cells with activated L1 jumping genes exhibit epigenetic instability, suggesting the critical role of epigenetic changes in regulating L1 jumping gene activity. Most of these epigenomic instabilities were found to arise during the early stages of embryogenesis. The study provides valuable insights into the aging process and the development of diseases in human colorectal tissues.
"This study illustrates that genomic damage in normal cells is acquired not only through exposure to carcinogens but also through the activity of endogenous components whose impact was previously unclear. Genomes of apparently healthy aged cells, particularly in the colorectal epithelium, become mosaic due to the activity of L1 jumping genes," said Prof. Young Seok Ju at KAIST.
"We emphasize the essential and ongoing collaboration among researchers in clinical medicine and basic medical sciences," said Prof. Min Jung Kim of the Department of Surgery at Seoul National University Hospital. "This case highlights the critical role of systematically collected human tissues from clinical settings in unraveling the complex process of disease development in humans."
"I am delighted that the research team's advancements in single-cell genome technology have come to fruition. We will persistently strive to lead in single-cell genome technology," said Prof. Hyun Woo Kwon of the Department of Nuclear Medicine at Korea University School of Medicine.
The research team received support from the Research Leader Program and the Young Researcher Program of the National Research Foundation of Korea, a grant from the MD-PhD/Medical Scientist Training Program through the Korea Health Industry Development Institute, and the Suh Kyungbae Foundation.
< Figure 1. Experimental design of the study >
< Figure 2. Schematic diagram illustrating factors influencing the soL1R landscape. >
Genetic composition of rc-L1s is inherited from the parents. The methylation landscape of rc-L1 promoters is predominantly determined by global DNA demethylation, followed by remethylation processes in the developmental stages. Then, when an rc-L1 is promoter demethylated in a specific cell lineage, the source expresses L1 transcripts thus making possible the induction of soL1Rs.
2023.05.22 View 9356
'Jumping Genes' Found to Alter Human Colon Genomes, Offering Insights into Aging and Tumorigenesis
The Korea Advanced Institute of Science and Technology (KAIST) and their collaborators have conducted a groundbreaking study targeting 'jumping genes' in the entire genomes of the human large intestine. Published in Nature on May 18 2023, the research unveils the surprising activity of 'Long interspersed nuclear element-1 (L1),' a type of jumping gene previously thought to be mostly dormant in human genomes. The study shows that L1 genes can become activated and disrupt genomic functions throughout an individual's lifetime, particularly in the colorectal epithelium.
(Paper Title: Widespread somatic L1 retrotransposition in normal colorectal epithelium, https://www.nature.com/articles/s41586-023-06046-z)
With approximately 500,000 L1 jumping genes, accounting for 17% of the human genome, they have long been recognized for their contribution to the evolution of the human species by introducing 'disruptive innovation' to genome sequences. Until now, it was believed that most L1 elements had lost their ability to jump in normal tissues of modern humans. However, this study reveals that some L1 jumping genes can be widely activated in normal cells, leading to the accumulation of genomic mutations over an individual's lifetime. The rate of L1 jumping and resulting genomic changes vary among different cell types, with a notable concentration observed in aged colon epithelial cells. The study illustrates that every colonic epithelial cell experiences an L1 jumping event by the age of 40 on average.
The research, led by co-first authors Chang Hyun Nam (a graduate student at KAIST) and Dr. Jeonghwan Youk (former graduate student at KAIST and assistant clinical professor at Seoul National University Hospital), involved the analysis of whole-genome sequences from 899 single cells obtained from skin (fibroblasts), blood, and colon epithelial tissues collected from 28 individuals. The study uncovers the activation of L1 jumping genes in normal cells, resulting in the gradual accumulation of genomic mutations over time. Additionally, the team explored epigenomic (DNA methylation) sequences to understand the mechanism behind L1 jumping gene activation. They found that cells with activated L1 jumping genes exhibit epigenetic instability, suggesting the critical role of epigenetic changes in regulating L1 jumping gene activity. Most of these epigenomic instabilities were found to arise during the early stages of embryogenesis. The study provides valuable insights into the aging process and the development of diseases in human colorectal tissues.
"This study illustrates that genomic damage in normal cells is acquired not only through exposure to carcinogens but also through the activity of endogenous components whose impact was previously unclear. Genomes of apparently healthy aged cells, particularly in the colorectal epithelium, become mosaic due to the activity of L1 jumping genes," said Prof. Young Seok Ju at KAIST.
"We emphasize the essential and ongoing collaboration among researchers in clinical medicine and basic medical sciences," said Prof. Min Jung Kim of the Department of Surgery at Seoul National University Hospital. "This case highlights the critical role of systematically collected human tissues from clinical settings in unraveling the complex process of disease development in humans."
"I am delighted that the research team's advancements in single-cell genome technology have come to fruition. We will persistently strive to lead in single-cell genome technology," said Prof. Hyun Woo Kwon of the Department of Nuclear Medicine at Korea University School of Medicine.
The research team received support from the Research Leader Program and the Young Researcher Program of the National Research Foundation of Korea, a grant from the MD-PhD/Medical Scientist Training Program through the Korea Health Industry Development Institute, and the Suh Kyungbae Foundation.
< Figure 1. Experimental design of the study >
< Figure 2. Schematic diagram illustrating factors influencing the soL1R landscape. >
Genetic composition of rc-L1s is inherited from the parents. The methylation landscape of rc-L1 promoters is predominantly determined by global DNA demethylation, followed by remethylation processes in the developmental stages. Then, when an rc-L1 is promoter demethylated in a specific cell lineage, the source expresses L1 transcripts thus making possible the induction of soL1Rs.
2023.05.22 View 9356 -
 Every Moment of Ultrafast Chemical Bonding Now Captured on Film
- The emerging moment of bond formation, two separate bonding steps, and subsequent vibrational motions were visualized. -
< Emergence of molecular vibrations and the evolution to covalent bonds observed in the research. Video Credit: KEK IMSS >
A team of South Korean researchers led by Professor Hyotcherl Ihee from the Department of Chemistry at KAIST reported the direct observation of the birthing moment of chemical bonds by tracking real-time atomic positions in the molecule. Professor Ihee, who also serves as Associate Director of the Center for Nanomaterials and Chemical Reactions at the Institute for Basic Science (IBS), conducted this study in collaboration with scientists at the Institute of Materials Structure Science of High Energy Accelerator Research Organization (KEK IMSS, Japan), RIKEN (Japan), and Pohang Accelerator Laboratory (PAL, South Korea). This work was published in Nature on June 24.
Targeted cancer drugs work by striking a tight bond between cancer cell and specific molecular targets that are involved in the growth and spread of cancer. Detailed images of such chemical bonding sites or pathways can provide key information necessary for maximizing the efficacy of oncogene treatments. However, atomic movements in a molecule have never been captured in the middle of the action, not even for an extremely simple molecule such as a triatomic molecule, made of only three atoms.
Professor Ihee's group and their international collaborators finally succeeded in capturing the ongoing reaction process of the chemical bond formation in the gold trimer. "The femtosecond-resolution images revealed that such molecular events took place in two separate stages, not simultaneously as previously assumed," says Professor Ihee, the corresponding author of the study. "The atoms in the gold trimer complex atoms remain in motion even after the chemical bonding is complete. The distance between the atoms increased and decreased periodically, exhibiting the molecular vibration. These visualized molecular vibrations allowed us to name the characteristic motion of each observed vibrational mode." adds Professor Ihee.
Atoms move extremely fast at a scale of femtosecond (fs) ― quadrillionths (or millionths of a billionth) of a second. Its movement is minute in the level of angstrom equal to one ten-billionth of a meter. They are especially elusive during the transition state where reaction intermediates are transitioning from reactants to products in a flash. The KAIST-IBS research team made this experimentally challenging task possible by using femtosecond x-ray liquidography (solution scattering). This experimental technique combines laser photolysis and x-ray scattering techniques. When a laser pulse strikes the sample, X-rays scatter and initiate the chemical bond formation reaction in the gold trimer complex. Femtosecond x-ray pulses obtained from a special light source called an x-ray free-electron laser (XFEL) were used to interrogate the bond-forming process. The experiments were performed at two XFEL facilities (4th generation linear accelerator) that are PAL-XFEL in South Korea and SACLA in Japan, and this study was conducted in collaboration with researchers from KEK IMSS, PAL, RIKEN, and the Japan Synchrotron Radiation Research Institute (JASRI).
Scattered waves from each atom interfere with each other and thus their x-ray scattering images are characterized by specific travel directions. The KAIST-IBS research team traced real-time positions of the three gold atoms over time by analyzing x-ray scattering images, which are determined by a three-dimensional structure of a molecule. Structural changes in the molecule complex resulted in multiple characteristic scattering images over time. When a molecule is excited by a laser pulse, multiple vibrational quantum states are simultaneously excited. The superposition of several excited vibrational quantum states is called a wave packet. The researchers tracked the wave packet in three-dimensional nuclear coordinates and found that the first half round of chemical bonding was formed within 35 fs after photoexcitation. The second half of the reaction followed within 360 fs to complete the entire reaction dynamics.
They also accurately illustrated molecular vibration motions in both temporal- and spatial-wise. This is quite a remarkable feat considering that such an ultrafast speed and a minute length of motion are quite challenging conditions for acquiring precise experimental data.
In this study, the KAIST-IBS research team improved upon their 2015 study published by Nature. In the previous study in 2015, the speed of the x-ray camera (time resolution) was limited to 500 fs, and the molecular structure had already changed to be linear with two chemical bonds within 500 fs. In this study, the progress of the bond formation and bent-to-linear structural transformation could be observed in real time, thanks to the improvement time resolution down to 100 fs. Thereby, the asynchronous bond formation mechanism in which two chemical bonds are formed in 35 fs and 360 fs, respectively, and the bent-to-linear transformation completed in 335 fs were visualized. In short, in addition to observing the beginning and end of chemical reactions, they reported every moment of the intermediate, ongoing rearrangement of nuclear configurations with dramatically improved experimental and analytical methods.
They will push this method of 'real-time tracking of atomic positions in a molecule and molecular vibration using femtosecond x-ray scattering' to reveal the mechanisms of organic and inorganic catalytic reactions and reactions involving proteins in the human body. "By directly tracking the molecular vibrations and real-time positions of all atoms in a molecule in the middle of reaction, we will be able to uncover mechanisms of various unknown organic and inorganic catalytic reactions and biochemical reactions," notes Dr. Jong Goo Kim, the lead author of the study.
Publications:
Kim, J. G., et al. (2020) ‘Mapping the emergence of molecular vibrations mediating bond formation’. Nature. Volume 582. Page 520-524. Available online at https://doi.org/10.1038/s41586-020-2417-3
Profile: Hyotcherl Ihee, Ph.D.
Professor
hyotcherl.ihee@kaist.ac.kr
http://time.kaist.ac.kr/
Ihee Laboratory
Department of Chemistry
KAIST
https://www.kaist.ac.kr
Daejeon 34141, Korea
(END)
2020.06.24 View 19711
Every Moment of Ultrafast Chemical Bonding Now Captured on Film
- The emerging moment of bond formation, two separate bonding steps, and subsequent vibrational motions were visualized. -
< Emergence of molecular vibrations and the evolution to covalent bonds observed in the research. Video Credit: KEK IMSS >
A team of South Korean researchers led by Professor Hyotcherl Ihee from the Department of Chemistry at KAIST reported the direct observation of the birthing moment of chemical bonds by tracking real-time atomic positions in the molecule. Professor Ihee, who also serves as Associate Director of the Center for Nanomaterials and Chemical Reactions at the Institute for Basic Science (IBS), conducted this study in collaboration with scientists at the Institute of Materials Structure Science of High Energy Accelerator Research Organization (KEK IMSS, Japan), RIKEN (Japan), and Pohang Accelerator Laboratory (PAL, South Korea). This work was published in Nature on June 24.
Targeted cancer drugs work by striking a tight bond between cancer cell and specific molecular targets that are involved in the growth and spread of cancer. Detailed images of such chemical bonding sites or pathways can provide key information necessary for maximizing the efficacy of oncogene treatments. However, atomic movements in a molecule have never been captured in the middle of the action, not even for an extremely simple molecule such as a triatomic molecule, made of only three atoms.
Professor Ihee's group and their international collaborators finally succeeded in capturing the ongoing reaction process of the chemical bond formation in the gold trimer. "The femtosecond-resolution images revealed that such molecular events took place in two separate stages, not simultaneously as previously assumed," says Professor Ihee, the corresponding author of the study. "The atoms in the gold trimer complex atoms remain in motion even after the chemical bonding is complete. The distance between the atoms increased and decreased periodically, exhibiting the molecular vibration. These visualized molecular vibrations allowed us to name the characteristic motion of each observed vibrational mode." adds Professor Ihee.
Atoms move extremely fast at a scale of femtosecond (fs) ― quadrillionths (or millionths of a billionth) of a second. Its movement is minute in the level of angstrom equal to one ten-billionth of a meter. They are especially elusive during the transition state where reaction intermediates are transitioning from reactants to products in a flash. The KAIST-IBS research team made this experimentally challenging task possible by using femtosecond x-ray liquidography (solution scattering). This experimental technique combines laser photolysis and x-ray scattering techniques. When a laser pulse strikes the sample, X-rays scatter and initiate the chemical bond formation reaction in the gold trimer complex. Femtosecond x-ray pulses obtained from a special light source called an x-ray free-electron laser (XFEL) were used to interrogate the bond-forming process. The experiments were performed at two XFEL facilities (4th generation linear accelerator) that are PAL-XFEL in South Korea and SACLA in Japan, and this study was conducted in collaboration with researchers from KEK IMSS, PAL, RIKEN, and the Japan Synchrotron Radiation Research Institute (JASRI).
Scattered waves from each atom interfere with each other and thus their x-ray scattering images are characterized by specific travel directions. The KAIST-IBS research team traced real-time positions of the three gold atoms over time by analyzing x-ray scattering images, which are determined by a three-dimensional structure of a molecule. Structural changes in the molecule complex resulted in multiple characteristic scattering images over time. When a molecule is excited by a laser pulse, multiple vibrational quantum states are simultaneously excited. The superposition of several excited vibrational quantum states is called a wave packet. The researchers tracked the wave packet in three-dimensional nuclear coordinates and found that the first half round of chemical bonding was formed within 35 fs after photoexcitation. The second half of the reaction followed within 360 fs to complete the entire reaction dynamics.
They also accurately illustrated molecular vibration motions in both temporal- and spatial-wise. This is quite a remarkable feat considering that such an ultrafast speed and a minute length of motion are quite challenging conditions for acquiring precise experimental data.
In this study, the KAIST-IBS research team improved upon their 2015 study published by Nature. In the previous study in 2015, the speed of the x-ray camera (time resolution) was limited to 500 fs, and the molecular structure had already changed to be linear with two chemical bonds within 500 fs. In this study, the progress of the bond formation and bent-to-linear structural transformation could be observed in real time, thanks to the improvement time resolution down to 100 fs. Thereby, the asynchronous bond formation mechanism in which two chemical bonds are formed in 35 fs and 360 fs, respectively, and the bent-to-linear transformation completed in 335 fs were visualized. In short, in addition to observing the beginning and end of chemical reactions, they reported every moment of the intermediate, ongoing rearrangement of nuclear configurations with dramatically improved experimental and analytical methods.
They will push this method of 'real-time tracking of atomic positions in a molecule and molecular vibration using femtosecond x-ray scattering' to reveal the mechanisms of organic and inorganic catalytic reactions and reactions involving proteins in the human body. "By directly tracking the molecular vibrations and real-time positions of all atoms in a molecule in the middle of reaction, we will be able to uncover mechanisms of various unknown organic and inorganic catalytic reactions and biochemical reactions," notes Dr. Jong Goo Kim, the lead author of the study.
Publications:
Kim, J. G., et al. (2020) ‘Mapping the emergence of molecular vibrations mediating bond formation’. Nature. Volume 582. Page 520-524. Available online at https://doi.org/10.1038/s41586-020-2417-3
Profile: Hyotcherl Ihee, Ph.D.
Professor
hyotcherl.ihee@kaist.ac.kr
http://time.kaist.ac.kr/
Ihee Laboratory
Department of Chemistry
KAIST
https://www.kaist.ac.kr
Daejeon 34141, Korea
(END)
2020.06.24 View 19711 -
 Scientists Discover the Mechanism of DNA High-Order Structure Formation
(Molecular structures of Abo1 in different energy states (left), Demonstration of an Abo1-assisted histone loading onto DNA by the DNA curtain assay. )
The genetic material of our cells—DNA—exists in a high-order structure called “chromatin”. Chromatin consists of DNA wrapped around histone proteins and efficiently packs DNA into a small volume. Moreover, using a spool and thread analogy, chromatin allows DNA to be locally wound or unwound, thus enabling genes to be enclosed or exposed. The misregulation of chromatin structures results in aberrant gene expression and can ultimately lead to developmental disorders or cancers. Despite the importance of DNA high-order structures, the complexity of the underlying machinery has circumvented molecular dissection.
For the first time, molecular biologists have uncovered how one particular mechanism uses energy to ensure proper histone placement onto DNA to form chromatin. They published their results on Dec. 17 in Nature Communications.
The study focused on proteins called histone chaperones. Histone chaperones are responsible for adding and removing specific histones at specific times during the DNA packaging process. The wrong histone at the wrong time and place could result in the misregulation of gene expression or aberrant DNA replication. Thus, histone chaperones are key players in the assembly and disassembly of chromatin.
“In order to carefully control the assembly and disassembly of chromatin units, histone chaperones act as molecular escorts that prevent histone aggregation and undesired interactions,” said Professor Ji-Joon Song in the Department of Biological Sciences at KAIST. “We set out to understand how a unique histone chaperone uses chemical energy to assemble or disassemble chromatin.”
Song and his team looked to Abo1, the only known histone chaperone that utilizes cellular energy (ATP). While Abo1 is found in yeast, it has an analogous partner in other organisms, including humans, called ATAD2. Both use ATP, which is produced through a cellular process where enzymes break down a molecule’s phosphate bond. ATP energy is typically used to power other cellular processes, but it is a rare partner for histone chaperones.
“This was an interesting problem in the field because all other histone chaperones studied to date do not use ATP,” Song said.
By imaging Abo1 with a single-molecule fluorescence imaging technique known as the DNA curtain assay, the researchers could examine the protein interactions at the single-molecule level. The technique allows scientists to arrange the DNA molecules and proteins on a single layer of a microfluidic chamber and examine the layer with fluorescence microscopy.
The researchers found through real-time observation that Abo1 is ring-shaped and changes its structure to accommodate a specific histone and deposit it on DNA. Moreover, they found that the accommodating structural changes are powered by ADP.
“We discovered a mechanism by which Abo1 accommodates histone substrates, ultimately allowing it to function as a unique energy-dependent histone chaperone,” Song said. “We also found that despite looking like a protein disassembly machine, Abo1 actually loads histone substrates onto DNA to facilitate chromatin assembly.”
The researchers plan to continue exploring how energy-dependent histone chaperones bind and release histones, with the ultimate goal of developing therapeutics that can target cancer-causing misbehavior by Abo1’s analogous human counterpart, ATAD2.
-Profile
Professor Ji-Joon Song
Department of Biological Sciences KI for the BioCentury (https://kis.kaist.ac.kr/index.php?mid=KIB_O) KAIST
2020.01.07 View 11804
Scientists Discover the Mechanism of DNA High-Order Structure Formation
(Molecular structures of Abo1 in different energy states (left), Demonstration of an Abo1-assisted histone loading onto DNA by the DNA curtain assay. )
The genetic material of our cells—DNA—exists in a high-order structure called “chromatin”. Chromatin consists of DNA wrapped around histone proteins and efficiently packs DNA into a small volume. Moreover, using a spool and thread analogy, chromatin allows DNA to be locally wound or unwound, thus enabling genes to be enclosed or exposed. The misregulation of chromatin structures results in aberrant gene expression and can ultimately lead to developmental disorders or cancers. Despite the importance of DNA high-order structures, the complexity of the underlying machinery has circumvented molecular dissection.
For the first time, molecular biologists have uncovered how one particular mechanism uses energy to ensure proper histone placement onto DNA to form chromatin. They published their results on Dec. 17 in Nature Communications.
The study focused on proteins called histone chaperones. Histone chaperones are responsible for adding and removing specific histones at specific times during the DNA packaging process. The wrong histone at the wrong time and place could result in the misregulation of gene expression or aberrant DNA replication. Thus, histone chaperones are key players in the assembly and disassembly of chromatin.
“In order to carefully control the assembly and disassembly of chromatin units, histone chaperones act as molecular escorts that prevent histone aggregation and undesired interactions,” said Professor Ji-Joon Song in the Department of Biological Sciences at KAIST. “We set out to understand how a unique histone chaperone uses chemical energy to assemble or disassemble chromatin.”
Song and his team looked to Abo1, the only known histone chaperone that utilizes cellular energy (ATP). While Abo1 is found in yeast, it has an analogous partner in other organisms, including humans, called ATAD2. Both use ATP, which is produced through a cellular process where enzymes break down a molecule’s phosphate bond. ATP energy is typically used to power other cellular processes, but it is a rare partner for histone chaperones.
“This was an interesting problem in the field because all other histone chaperones studied to date do not use ATP,” Song said.
By imaging Abo1 with a single-molecule fluorescence imaging technique known as the DNA curtain assay, the researchers could examine the protein interactions at the single-molecule level. The technique allows scientists to arrange the DNA molecules and proteins on a single layer of a microfluidic chamber and examine the layer with fluorescence microscopy.
The researchers found through real-time observation that Abo1 is ring-shaped and changes its structure to accommodate a specific histone and deposit it on DNA. Moreover, they found that the accommodating structural changes are powered by ADP.
“We discovered a mechanism by which Abo1 accommodates histone substrates, ultimately allowing it to function as a unique energy-dependent histone chaperone,” Song said. “We also found that despite looking like a protein disassembly machine, Abo1 actually loads histone substrates onto DNA to facilitate chromatin assembly.”
The researchers plan to continue exploring how energy-dependent histone chaperones bind and release histones, with the ultimate goal of developing therapeutics that can target cancer-causing misbehavior by Abo1’s analogous human counterpart, ATAD2.
-Profile
Professor Ji-Joon Song
Department of Biological Sciences KI for the BioCentury (https://kis.kaist.ac.kr/index.php?mid=KIB_O) KAIST
2020.01.07 View 11804 -
 Two Professors Recognized for the National R&D Excellence 100
< Professor Haeng-Ki Lee (left) and Professor Jeong-Ho Lee (right) >
Two KAIST professors were listed among the 2019 National R&D Excellence 100 announced by the Ministry of Science and ICT and the Korea Institute of S&T Evaluation and Planning.
Professor Haeng-Ki Lee from the Department of Civil and Environmental Engineering was recognized in the field of mechanics and materials for his research on developing new construction materials through the convergence of nano- and biotechnologies.
In the field of life and marine science, Professor Jeong-Ho Lee from the Graduate School of Medical Science and Engineering was lauded for his research of diagnostic tools and therapies for glioblastoma and pediatric brain tumors.
A certificate from the Minister of Ministry of Science and ICT will be conferred to these two professors, and their names will be inscribed on a special 2019 National R&D Excellence 100 plaque to celebrate their achievements. The professors will also be given privileges during the process of new R&D project selection.
(END)
2019.10.15 View 13717
Two Professors Recognized for the National R&D Excellence 100
< Professor Haeng-Ki Lee (left) and Professor Jeong-Ho Lee (right) >
Two KAIST professors were listed among the 2019 National R&D Excellence 100 announced by the Ministry of Science and ICT and the Korea Institute of S&T Evaluation and Planning.
Professor Haeng-Ki Lee from the Department of Civil and Environmental Engineering was recognized in the field of mechanics and materials for his research on developing new construction materials through the convergence of nano- and biotechnologies.
In the field of life and marine science, Professor Jeong-Ho Lee from the Graduate School of Medical Science and Engineering was lauded for his research of diagnostic tools and therapies for glioblastoma and pediatric brain tumors.
A certificate from the Minister of Ministry of Science and ICT will be conferred to these two professors, and their names will be inscribed on a special 2019 National R&D Excellence 100 plaque to celebrate their achievements. The professors will also be given privileges during the process of new R&D project selection.
(END)
2019.10.15 View 13717 -
 Early Genome Catastrophes Can Cause Non-Smoking Lung Cancer
Some teenagers harbor catastrophic changes to their genomes that can lead to lung cancer later on in life, even if they never smoke
(Professor Young Seok Ju at the Graduate School of Medical Science and Engineering)
Catastrophic rearrangements in the genome occurring as early as childhood and adolescence can lead to the development of lung cancer in later years in non-smokers. This finding, published in Cell, helps explain how some non-smoking-related lung cancers develop.
Researchers at KAIST, Seoul National University and their collaborators confirmed that gene fusions in non-smokers mostly occur early on, sometimes as early as childhood or adolescence, and on average about three decades before cancer is diagnosed. The study showed that these mutant lung cells, harboring oncogenic seeds, remain dormant for several decades until a number of further mutations accumulate sufficiently for progression into cancer. This is the first study to reveal the landscape of genome structural variations in lung adenocarcinoma.
Lung cancer is the leading cause of cancer-related deaths worldwide, and lung adenocarcinoma is its most common type. Most lung adenocarcinomas are associated with chronic smoking, but about a fourth develop in non-smokers. Precisely what happens in non-smokers for this cancer to develop is not clearly understood.
Researchers analyzed the genomes of 138 lung adenocarcinoma patients, including smokers and non-smokers, with whole-genome sequencing technologies. They explored DNA damage that induced neoplastic transformation.
Lung adenocarcinomas that originated from chronic smoking, referred to as signature 4-high (S4-high) cancers in the study, showed several distinguishing features compared to smoking-unrelated cancers (S4-low).
People in the S4-high group were largely older, men and had more frequent mutations in a cancer-related gene called KRAS. Cancer genomes in the S4-high group were hypermutated with simple mutational classes, such as the substitution, insertion, or deletion of a single base, the building block of DNA.
But the story was very different in the S4-low group. Generally, mutational profiles in this group were much more silent than the S4-high group. However, all cancer-related gene fusions, which are abnormally activated from the merging of two originally separate genes, were exclusively observed in the S4-low group.
The patterns of genomic structural changes underlying gene fusions suggest that about three in four cases of gene fusions emerged from a single cellular crisis causing massive genomic fragmentation and subsequent imprecise repair in normal lung epithelium.
Most strikingly, these major genomic rearrangements, which led to the development of lung adenocarcinoma, are very likely to be acquired decades before cancer diagnosis. The researchers used genomic archaeology techniques to trace the timing of when the catastrophes took place.
Researchers started this study seven years ago when they discovered the expression of the KIF5B-RET gene fusion in lung adenocarcinoma for the first time. Professor Young-Seok Ju, co-lead author from the Graduate School of Medical Science and Engineering at KAIST says, “It is remarkable that oncogenesis can begin by a massive shattering of chromosomes early in life. Our study immediately raises a new question: What induces the mutational catastrophe in our normal lung epithelium.”
Professor Young Tae Kim, co-lead author from Seoul National University says, “We hope this work will help us get one step closer to precision medicine for lung cancer patients.”
The research team plans to further focus on the molecular mechanisms that stimulate complex rearrangements in the body, through screening the genomic structures of fusion genes in other cancer types.
This study was supported by the National Research Foundation of Korea (NRF), Korea Health Industry Development Institute (KHIDI), Suh Kyungbae Foundation, the College of Medicine Research Foundations at Seoul National University and others.
Figure.
(Smoking-unrelated oncogenesis of lung cancers by gene fusions)
Publication.
Jake June-Koo Lee, Seongyeol Park et al., Tracing Oncogene Rearrangements in the Mutational History of Lung Adenocarcinoma
Cell 177, June 13 2019, online publication ahead of print at May 30, 2019
https://doi.org/10.1016/j.cell.2019.05.013
Profile: Prof Young Seok Ju, MD, PhD
ysju@kaist.ac.kr
http://julab.kaist.ac.kr
Associate Professor
Graduate School of Medical Science and Engineering (GSMSE)
Korea Advanced Institute of Science and Technology (KAIST)
Daejeon 34141, Korea
Profile: Prof Young Tae Kim, MD, PhD
ytkim@snu.ac.kr
Professor
Seoul National University Cancer Research Institute
Department of Thoracic and Cardiovascular Surgery
Seoul National University Hospital Seoul 03080, Korea
2019.05.31 View 58358
Early Genome Catastrophes Can Cause Non-Smoking Lung Cancer
Some teenagers harbor catastrophic changes to their genomes that can lead to lung cancer later on in life, even if they never smoke
(Professor Young Seok Ju at the Graduate School of Medical Science and Engineering)
Catastrophic rearrangements in the genome occurring as early as childhood and adolescence can lead to the development of lung cancer in later years in non-smokers. This finding, published in Cell, helps explain how some non-smoking-related lung cancers develop.
Researchers at KAIST, Seoul National University and their collaborators confirmed that gene fusions in non-smokers mostly occur early on, sometimes as early as childhood or adolescence, and on average about three decades before cancer is diagnosed. The study showed that these mutant lung cells, harboring oncogenic seeds, remain dormant for several decades until a number of further mutations accumulate sufficiently for progression into cancer. This is the first study to reveal the landscape of genome structural variations in lung adenocarcinoma.
Lung cancer is the leading cause of cancer-related deaths worldwide, and lung adenocarcinoma is its most common type. Most lung adenocarcinomas are associated with chronic smoking, but about a fourth develop in non-smokers. Precisely what happens in non-smokers for this cancer to develop is not clearly understood.
Researchers analyzed the genomes of 138 lung adenocarcinoma patients, including smokers and non-smokers, with whole-genome sequencing technologies. They explored DNA damage that induced neoplastic transformation.
Lung adenocarcinomas that originated from chronic smoking, referred to as signature 4-high (S4-high) cancers in the study, showed several distinguishing features compared to smoking-unrelated cancers (S4-low).
People in the S4-high group were largely older, men and had more frequent mutations in a cancer-related gene called KRAS. Cancer genomes in the S4-high group were hypermutated with simple mutational classes, such as the substitution, insertion, or deletion of a single base, the building block of DNA.
But the story was very different in the S4-low group. Generally, mutational profiles in this group were much more silent than the S4-high group. However, all cancer-related gene fusions, which are abnormally activated from the merging of two originally separate genes, were exclusively observed in the S4-low group.
The patterns of genomic structural changes underlying gene fusions suggest that about three in four cases of gene fusions emerged from a single cellular crisis causing massive genomic fragmentation and subsequent imprecise repair in normal lung epithelium.
Most strikingly, these major genomic rearrangements, which led to the development of lung adenocarcinoma, are very likely to be acquired decades before cancer diagnosis. The researchers used genomic archaeology techniques to trace the timing of when the catastrophes took place.
Researchers started this study seven years ago when they discovered the expression of the KIF5B-RET gene fusion in lung adenocarcinoma for the first time. Professor Young-Seok Ju, co-lead author from the Graduate School of Medical Science and Engineering at KAIST says, “It is remarkable that oncogenesis can begin by a massive shattering of chromosomes early in life. Our study immediately raises a new question: What induces the mutational catastrophe in our normal lung epithelium.”
Professor Young Tae Kim, co-lead author from Seoul National University says, “We hope this work will help us get one step closer to precision medicine for lung cancer patients.”
The research team plans to further focus on the molecular mechanisms that stimulate complex rearrangements in the body, through screening the genomic structures of fusion genes in other cancer types.
This study was supported by the National Research Foundation of Korea (NRF), Korea Health Industry Development Institute (KHIDI), Suh Kyungbae Foundation, the College of Medicine Research Foundations at Seoul National University and others.
Figure.
(Smoking-unrelated oncogenesis of lung cancers by gene fusions)
Publication.
Jake June-Koo Lee, Seongyeol Park et al., Tracing Oncogene Rearrangements in the Mutational History of Lung Adenocarcinoma
Cell 177, June 13 2019, online publication ahead of print at May 30, 2019
https://doi.org/10.1016/j.cell.2019.05.013
Profile: Prof Young Seok Ju, MD, PhD
ysju@kaist.ac.kr
http://julab.kaist.ac.kr
Associate Professor
Graduate School of Medical Science and Engineering (GSMSE)
Korea Advanced Institute of Science and Technology (KAIST)
Daejeon 34141, Korea
Profile: Prof Young Tae Kim, MD, PhD
ytkim@snu.ac.kr
Professor
Seoul National University Cancer Research Institute
Department of Thoracic and Cardiovascular Surgery
Seoul National University Hospital Seoul 03080, Korea
2019.05.31 View 58358 -
 Special Lecture by Professor Sung-Hou Kim of UC Berkeley
As part of its special lecture series, the Department of Biological Sciences at KAIST has invited Professor Sung-Hou Kim of the Department of Chemistry at the University of California, Berkeley, to lecture on his research in structural biology. He will speak twice on May 23 and 30, respectively, on the topics “Origin of Universe and Earth—A Narrative” and “Origin of Life and Human Species—A Narrative.”
Professor Kim's research addresses the structural basis of molecules to reveal how they communicate with each other to activate or inhibit particular processes in cell growth, cell differentiation, and cancer. Using the single-crystal X-ray diffraction technology, he discovered, for the first time in the world, the three-dimensional (3-D) structure of a transfer RNA (t-RNA) and received much praise for this work from the scientific community. Since then, he has been cited as a candidate for a Nobel Prize in Chemistry for many years.
He also examined the 3-D structures of a RAS protein in normal and cancer cells and identified the mutations of the RAS protein as a cause for cancer. His work has assisted in the development of target drugs for cancer treatment. In recent years, he has adopted a computational biology approach to study the structure and function of biological genomics, with which he has tried to predict disease-sensitive genes.
Professor Kim graduated from Seoul National University in 1962 and received his Ph.D. degree in chemistry from the University of Pittsburgh in the United States in 1966. He worked at the Massachusetts Institute of Technology (MIT) as a senior research scientist, and has taught at UC Berkeley since 1978.
2016.05.23 View 7761
Special Lecture by Professor Sung-Hou Kim of UC Berkeley
As part of its special lecture series, the Department of Biological Sciences at KAIST has invited Professor Sung-Hou Kim of the Department of Chemistry at the University of California, Berkeley, to lecture on his research in structural biology. He will speak twice on May 23 and 30, respectively, on the topics “Origin of Universe and Earth—A Narrative” and “Origin of Life and Human Species—A Narrative.”
Professor Kim's research addresses the structural basis of molecules to reveal how they communicate with each other to activate or inhibit particular processes in cell growth, cell differentiation, and cancer. Using the single-crystal X-ray diffraction technology, he discovered, for the first time in the world, the three-dimensional (3-D) structure of a transfer RNA (t-RNA) and received much praise for this work from the scientific community. Since then, he has been cited as a candidate for a Nobel Prize in Chemistry for many years.
He also examined the 3-D structures of a RAS protein in normal and cancer cells and identified the mutations of the RAS protein as a cause for cancer. His work has assisted in the development of target drugs for cancer treatment. In recent years, he has adopted a computational biology approach to study the structure and function of biological genomics, with which he has tried to predict disease-sensitive genes.
Professor Kim graduated from Seoul National University in 1962 and received his Ph.D. degree in chemistry from the University of Pittsburgh in the United States in 1966. He worked at the Massachusetts Institute of Technology (MIT) as a senior research scientist, and has taught at UC Berkeley since 1978.
2016.05.23 View 7761 -
 Professor Suk-Joong Kang Receives the Richard Brook and Helmholtz Awards
Professor Suk-Joong Kang of KAIST’s Department of Materials Sciences and Engineering received the Richard Brook Award from the European Ceramic Society at its 14th conference held on June 21, 2015, in Toledo, Spain. The award is presented to the most distinguished academic or engineer in ceramics from a non-European country. Professor Kang gave the commemorative lecture after the award ceremony.
Professor Kang is an expert in the field of sintering and microstructural evolution in ceramics and metals. He suggested a new model for grain growth and identified the principles of microstructural evolution.
He also received the 2015 Helmholtz Fellow Award in June. The Helmholtz Association, the largest scientific organization in Germany, confers the award on outstanding senior scientists based outside Germany who have made great academic and research achievements in their fields.
Professor Kang said of the Brook Award, “It is such an honor to receive an award from an eminent global institution. I take this opportunity to thank my students and colleagues for their support, and I will work harder for my research.”
2015.07.20 View 7484
Professor Suk-Joong Kang Receives the Richard Brook and Helmholtz Awards
Professor Suk-Joong Kang of KAIST’s Department of Materials Sciences and Engineering received the Richard Brook Award from the European Ceramic Society at its 14th conference held on June 21, 2015, in Toledo, Spain. The award is presented to the most distinguished academic or engineer in ceramics from a non-European country. Professor Kang gave the commemorative lecture after the award ceremony.
Professor Kang is an expert in the field of sintering and microstructural evolution in ceramics and metals. He suggested a new model for grain growth and identified the principles of microstructural evolution.
He also received the 2015 Helmholtz Fellow Award in June. The Helmholtz Association, the largest scientific organization in Germany, confers the award on outstanding senior scientists based outside Germany who have made great academic and research achievements in their fields.
Professor Kang said of the Brook Award, “It is such an honor to receive an award from an eminent global institution. I take this opportunity to thank my students and colleagues for their support, and I will work harder for my research.”
2015.07.20 View 7484 -
 Science Daily: Nanostructured Capsules Could Bring About Paints and Electronic Displays That Never Fade
A collaborative research by Professor Shin-Hyun Kim of Chemical and Bimolecular Engineering, KAIST, and his student, Tae-Min Choi, on nano-structural colors with Harvard University was published by Science Daily on March 14, 2014.
For the article, please go to:
Science Daily,
March 14, 2014
Featured Research
"Brighter inks, without pigment: Nanostructured capsules could bring about paints and electronic displays that never fade"
http://www.sciencedaily.com/releases/2014/03/140314164214.htm
2014.03.17 View 10497
Science Daily: Nanostructured Capsules Could Bring About Paints and Electronic Displays That Never Fade
A collaborative research by Professor Shin-Hyun Kim of Chemical and Bimolecular Engineering, KAIST, and his student, Tae-Min Choi, on nano-structural colors with Harvard University was published by Science Daily on March 14, 2014.
For the article, please go to:
Science Daily,
March 14, 2014
Featured Research
"Brighter inks, without pigment: Nanostructured capsules could bring about paints and electronic displays that never fade"
http://www.sciencedaily.com/releases/2014/03/140314164214.htm
2014.03.17 View 10497 -
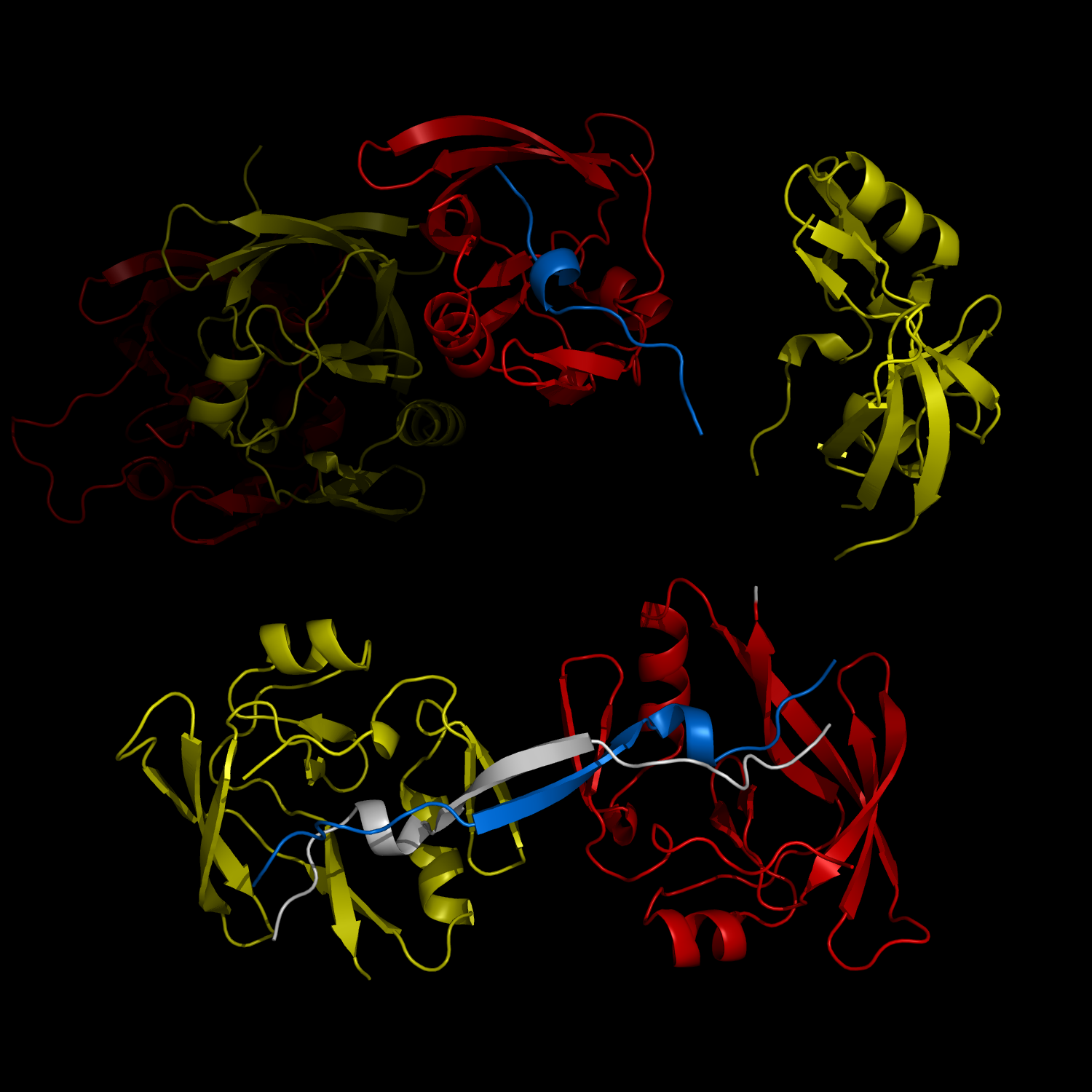 New Structural Insight into Neurodegenerative Disease
A research team from the Korea Advanced Institute of Science and Technology (KAIST) released their results on the structure and molecular details of the neurodegenerative disease-associated protein Ataxin-1. Mutations in Ataxin-1 cause the neurological disease, Spinocerebella Ataxia Type 1 (SCA1), which is characterized by a loss of muscular coordination and balance (ataxia), as is seen in Parkinson’s, Alzheimer’s, and Huntington’s diseases.
SCA1-causing mutations in the ATAXIN1 gene alter the length of a glutamine stretch in the Ataxin-1 protein. The research team provides the first structural insight into the complex formation of ATAXIN-1 with its binding partner, Capicua (CIC). The team, led by Professor Ji-Joon Song from the Department of Biological Sciences at KAIST, solved the structure of Ataxin-1 and CIC complex in atomic level revealing molecular details of the interaction between Ataxin-1 and CIC.
Professor Song explained his recent research work,
“We are able to see the intricate process of complex formation and reconfiguration of the two proteins when they interact with each other. Our work, we expect, will provide a new therapeutic target to modulate SCA1 neurodegenerative disease.”
Understanding structural and molecular details of proteins at the atomic level will help researchers to track the molecular pathogenesis of the disease and, ultimately, design targeted therapies or treatments for patients, rather than just relieving the symptoms of diseases.
Professor Song’s research paper, entitled “Structural Basis of Protein Complex Formation and Reconfiguration by Polyglutamine Disease Protein ATAXIN-1 and Capicua,” will be published in the March 15th issue of Genes & Development (www.genesdev.org).
Complex Formation and Reconfiguration of ATAXIN-1 and Capicua
The complex formation between a polyglutamine disease protein, ATXIN-1 and the transcriptional repressor Capicua (CIC) plays a critical role in SCA 1 pathogenesis. The image shows that the homodimerization of ATXIN-1 (yellow and red) is disrupted upon binding of CIC (blue). Furthermore, the binding of CIC to the ATXIN-1 induces a new form of ATXIN-1 dimerization mediated by CICs (ATXIN-1 AXH domains are shown in yellow and red, and CIC peptides shown in blue and white).
2013.04.02 View 10209
New Structural Insight into Neurodegenerative Disease
A research team from the Korea Advanced Institute of Science and Technology (KAIST) released their results on the structure and molecular details of the neurodegenerative disease-associated protein Ataxin-1. Mutations in Ataxin-1 cause the neurological disease, Spinocerebella Ataxia Type 1 (SCA1), which is characterized by a loss of muscular coordination and balance (ataxia), as is seen in Parkinson’s, Alzheimer’s, and Huntington’s diseases.
SCA1-causing mutations in the ATAXIN1 gene alter the length of a glutamine stretch in the Ataxin-1 protein. The research team provides the first structural insight into the complex formation of ATAXIN-1 with its binding partner, Capicua (CIC). The team, led by Professor Ji-Joon Song from the Department of Biological Sciences at KAIST, solved the structure of Ataxin-1 and CIC complex in atomic level revealing molecular details of the interaction between Ataxin-1 and CIC.
Professor Song explained his recent research work,
“We are able to see the intricate process of complex formation and reconfiguration of the two proteins when they interact with each other. Our work, we expect, will provide a new therapeutic target to modulate SCA1 neurodegenerative disease.”
Understanding structural and molecular details of proteins at the atomic level will help researchers to track the molecular pathogenesis of the disease and, ultimately, design targeted therapies or treatments for patients, rather than just relieving the symptoms of diseases.
Professor Song’s research paper, entitled “Structural Basis of Protein Complex Formation and Reconfiguration by Polyglutamine Disease Protein ATAXIN-1 and Capicua,” will be published in the March 15th issue of Genes & Development (www.genesdev.org).
Complex Formation and Reconfiguration of ATAXIN-1 and Capicua
The complex formation between a polyglutamine disease protein, ATXIN-1 and the transcriptional repressor Capicua (CIC) plays a critical role in SCA 1 pathogenesis. The image shows that the homodimerization of ATXIN-1 (yellow and red) is disrupted upon binding of CIC (blue). Furthermore, the binding of CIC to the ATXIN-1 induces a new form of ATXIN-1 dimerization mediated by CICs (ATXIN-1 AXH domains are shown in yellow and red, and CIC peptides shown in blue and white).
2013.04.02 View 10209 -
 Biomimetic reflective display technology developed
Professor Shin Jung Hoon
The bright colors of a rainbow or a peacock are produced by the reflection and interference of light in transparent periodic structures, producing what is called a structural color. These colors are very bright and change according to the viewing angle. On the other hand, the wings of a morpho-butterfly also have structural colors but are predominantly blue over a wide range of angles. This is because the unique structure of the morpho-butterfly’s wings contains both order and chaos.
Professor Shin Jung Hoon’s team from the Department of Physics and the Graduate School of Nanoscience and Technology at KAIST produced a display that mimics the structure of the morpho-butterfly’s wings using glass beads.
This research successfully produced a reflective display (one that reflects external light to project images), which could be used to make very bright displays with low energy consumption. This technology can also be used to make anti-counterfeit bills, as well as coating materials for mobile phones and wallets.
The structure of the morpho-butterfly’s wings seems to be in periodic order at the 1-micrometer level, but contains disorder at the 100-nanometer level. So far, no one had succeeded in reproducing a structure with both order and disorder at the nanometer level.
Professor Shin’s team randomly aligned differently sized glass beads of a few hundred nanometers to create chaos and placed a thin periodic film on top of it using the semiconductor deposition method, thereby creating the morpho-butterfly-like structure over a large area.
This new development produced better color and brightness than the morpho-butterfly wing and even exhibited less color change according to angle. The team sealed the film in thin plastic, which helped to maintain the superior properties whilst making it more firm and paper-like.
Professor Shin emphasized that the results were an exemplary success in the field of biomimetics and that structural colors could have other applications in sensors and fashion, for example.
The results were first introduced on May 3rd in Nature as one of the Research Highlights and will be published in the online version of the material science magazine, Advanced Materials.
This research was jointly conducted by Professor Shin Jung Hoon (Department of Physics / Graduate School of Nanoscience and Technology at KAIST), Professor Park NamKyoo (Department of Electrical and Computer Engineering at Seoul National University), and Samsung Advanced Institute of Technology. The funding was provided by the National Research Foundation of Korea and the Ministry of Education, Science and Technology as part of the World Class University (WCU) project.
Figure 2. The biomimetic film can express many different colors
Figure 3. The biomimetic diplay and a morpho-butterfly
2012.05.07 View 16766
Biomimetic reflective display technology developed
Professor Shin Jung Hoon
The bright colors of a rainbow or a peacock are produced by the reflection and interference of light in transparent periodic structures, producing what is called a structural color. These colors are very bright and change according to the viewing angle. On the other hand, the wings of a morpho-butterfly also have structural colors but are predominantly blue over a wide range of angles. This is because the unique structure of the morpho-butterfly’s wings contains both order and chaos.
Professor Shin Jung Hoon’s team from the Department of Physics and the Graduate School of Nanoscience and Technology at KAIST produced a display that mimics the structure of the morpho-butterfly’s wings using glass beads.
This research successfully produced a reflective display (one that reflects external light to project images), which could be used to make very bright displays with low energy consumption. This technology can also be used to make anti-counterfeit bills, as well as coating materials for mobile phones and wallets.
The structure of the morpho-butterfly’s wings seems to be in periodic order at the 1-micrometer level, but contains disorder at the 100-nanometer level. So far, no one had succeeded in reproducing a structure with both order and disorder at the nanometer level.
Professor Shin’s team randomly aligned differently sized glass beads of a few hundred nanometers to create chaos and placed a thin periodic film on top of it using the semiconductor deposition method, thereby creating the morpho-butterfly-like structure over a large area.
This new development produced better color and brightness than the morpho-butterfly wing and even exhibited less color change according to angle. The team sealed the film in thin plastic, which helped to maintain the superior properties whilst making it more firm and paper-like.
Professor Shin emphasized that the results were an exemplary success in the field of biomimetics and that structural colors could have other applications in sensors and fashion, for example.
The results were first introduced on May 3rd in Nature as one of the Research Highlights and will be published in the online version of the material science magazine, Advanced Materials.
This research was jointly conducted by Professor Shin Jung Hoon (Department of Physics / Graduate School of Nanoscience and Technology at KAIST), Professor Park NamKyoo (Department of Electrical and Computer Engineering at Seoul National University), and Samsung Advanced Institute of Technology. The funding was provided by the National Research Foundation of Korea and the Ministry of Education, Science and Technology as part of the World Class University (WCU) project.
Figure 2. The biomimetic film can express many different colors
Figure 3. The biomimetic diplay and a morpho-butterfly
2012.05.07 View 16766