Tae-Young+Yoon
-
 Mapping the Folding Process of a Single Membrane Protein
KAIST and UCLA scientists were able to observe an individual membrane protein fold and unfold by pulling and releasing magnetically trapped protein molecules.
Proteins are huge molecules containing hundreds to thousands of atoms that adopt a unique three dimensional structure, placing chemical groups in just the right place to catalyze reactions or build cellular structures. How all those atoms manage to find the right location - the so-called folding problem - has fascinated molecular biologists since the first structures were seen in the 1950s. Moreover, folding has important medical implications because most genetic defects cause protein misfolding.
About a third of all proteins float around in the cell membrane where they ensure the right chemicals get in the cell in the right amounts. Membrane proteins also provide key information links between the cell and its environment. Indeed, most drugs target membrane proteins. Nevertheless, the folding of membrane proteins has been particularly difficult to study and has rarely been studied in natural environments, leaving the folding process for a large fraction of the protein universe still largely cloaked in mystery.
In a recent issue of Nature Chemical Biology, published on October 19, 2015, a research team led by Tae-Young Yoon of the Department of Physics at the Korea Advanced Institute of Science and Technology (KAIST) and James U. Bowie of the Department of Chemistry and Biochemistry at the University of California, Los Angeles (UCLA), report a new method for manipulating the folding of membrane proteins in a membrane environment using a tool called a magnetic tweezer.
Researchers first attach long DNA handles to the ends of the protein. One handle is attached to a glass surface and the other to a magnetic bead. Using a magnet, they can essentially grab the protein and pull on it, inducing it to unfold. By playing with the bead attached to the protein, they can force the protein to unfold or allow it to refold, and watch all this happening by 3D-tracking of the magnetic bead. With this novel strategy, they were able to quantitatively map the folding energy landscape, the folding kinetic rate, and folding intermediates of a membrane protein in a membrane environment for the first time.
“I have been dreaming about this experiment for a decade. To see it work so well is really gratifying,” said Dr. Bowie.
One of the major surprises in the study was that essentially all the atoms of the protein jump into the correct structure together. The researchers expected that the protein structure would come together in a more piecemeal fashion, with different parts of the structure forming separately, but that was not the case. It is possible that nature evolved such a smooth, highly cooperative folding process to prevent partially folded forms that could get into trouble in the crowded cell membrane. On the other hand, the cooperative folding seen here might not apply to other membrane proteins.
“We need to look at more proteins. The technique developed here may allow us to do just that,” said Dr. Yoon.
The single molecule mechanical manipulation technique could enable detailed folding studies of many other membrane proteins. A major barrier to the study of membrane proteins previously is that the proteins tend to stick together and get tangled up, as computer cords lying at your feet tend to do. With the tweezer technique used in this work, the protein cords are held apart from other cords so they can’t get knotted up. It is hoped that the new approach will open up an important part of the protein universe to scrutiny, including many proteins that become misfolded in disease states.
The title of the research paper is “Mapping the energy landscape for second-stage folding of a single membrane protein” (DOI: 10.1038/nchembio.1939).
Picture: Single-molecule magnetic tweezers that induce mechanical unfolding and refolding of a single membrane protein. Since the force applied is parallel to the biological lipid membrane, the unfolding and refolding processes occur within the membrane.
2015.10.20 View 10698
Mapping the Folding Process of a Single Membrane Protein
KAIST and UCLA scientists were able to observe an individual membrane protein fold and unfold by pulling and releasing magnetically trapped protein molecules.
Proteins are huge molecules containing hundreds to thousands of atoms that adopt a unique three dimensional structure, placing chemical groups in just the right place to catalyze reactions or build cellular structures. How all those atoms manage to find the right location - the so-called folding problem - has fascinated molecular biologists since the first structures were seen in the 1950s. Moreover, folding has important medical implications because most genetic defects cause protein misfolding.
About a third of all proteins float around in the cell membrane where they ensure the right chemicals get in the cell in the right amounts. Membrane proteins also provide key information links between the cell and its environment. Indeed, most drugs target membrane proteins. Nevertheless, the folding of membrane proteins has been particularly difficult to study and has rarely been studied in natural environments, leaving the folding process for a large fraction of the protein universe still largely cloaked in mystery.
In a recent issue of Nature Chemical Biology, published on October 19, 2015, a research team led by Tae-Young Yoon of the Department of Physics at the Korea Advanced Institute of Science and Technology (KAIST) and James U. Bowie of the Department of Chemistry and Biochemistry at the University of California, Los Angeles (UCLA), report a new method for manipulating the folding of membrane proteins in a membrane environment using a tool called a magnetic tweezer.
Researchers first attach long DNA handles to the ends of the protein. One handle is attached to a glass surface and the other to a magnetic bead. Using a magnet, they can essentially grab the protein and pull on it, inducing it to unfold. By playing with the bead attached to the protein, they can force the protein to unfold or allow it to refold, and watch all this happening by 3D-tracking of the magnetic bead. With this novel strategy, they were able to quantitatively map the folding energy landscape, the folding kinetic rate, and folding intermediates of a membrane protein in a membrane environment for the first time.
“I have been dreaming about this experiment for a decade. To see it work so well is really gratifying,” said Dr. Bowie.
One of the major surprises in the study was that essentially all the atoms of the protein jump into the correct structure together. The researchers expected that the protein structure would come together in a more piecemeal fashion, with different parts of the structure forming separately, but that was not the case. It is possible that nature evolved such a smooth, highly cooperative folding process to prevent partially folded forms that could get into trouble in the crowded cell membrane. On the other hand, the cooperative folding seen here might not apply to other membrane proteins.
“We need to look at more proteins. The technique developed here may allow us to do just that,” said Dr. Yoon.
The single molecule mechanical manipulation technique could enable detailed folding studies of many other membrane proteins. A major barrier to the study of membrane proteins previously is that the proteins tend to stick together and get tangled up, as computer cords lying at your feet tend to do. With the tweezer technique used in this work, the protein cords are held apart from other cords so they can’t get knotted up. It is hoped that the new approach will open up an important part of the protein universe to scrutiny, including many proteins that become misfolded in disease states.
The title of the research paper is “Mapping the energy landscape for second-stage folding of a single membrane protein” (DOI: 10.1038/nchembio.1939).
Picture: Single-molecule magnetic tweezers that induce mechanical unfolding and refolding of a single membrane protein. Since the force applied is parallel to the biological lipid membrane, the unfolding and refolding processes occur within the membrane.
2015.10.20 View 10698 -
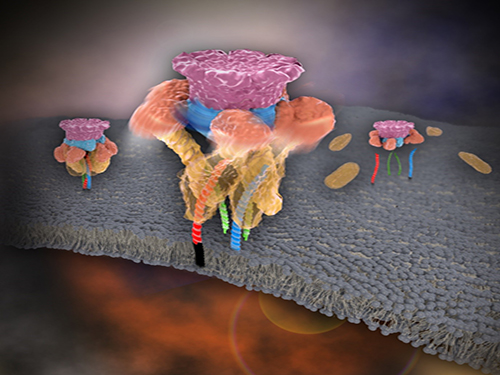 Mystery in Membrane Traffic How NSF Disassembles Single SNAR Complex Solved
KAIST researchers discovered that the protein N-ethylmaleimide-sensitive factor (NSF) unravels a single SNARE complex using one round ATP turnover by tearing the complex with a single burst, contradicting a previous theory that it unwinds in a processive manner.
In 2013, James E. Rothman, Randy W. Schekman, and Thomas C. Südhof won the Nobel Prize in Physiology or Medicine for their discoveries of molecular machineries for vesicle trafficking, a major transport system in cells for maintaining cellular processes. Vesicle traffic acts as a kind of “home-delivery service” in cells. Vesicles package and deliver materials such as proteins and hormones from one cell organelle to another. Then it releases its contents by fusing with the target organelle’s membrane. One example of vesicle traffic is in neuronal communications, where neurotransmitters are released from a neuron. Some of the key proteins for vesicle traffic discovered by the Nobel Prize winners were N-ethylmaleimide-sensitive factor (NSF), alpha-soluble NSF attachment protein (α-SNAP), and soluble SNAP receptors (SNAREs).
SNARE proteins are known as the minimal machinery for membrane fusion. To induce membrane fusion, the proteins combine to form a SNARE complex in a four helical bundle, and NSF and α-SNAP disassemble the SNARE complex for reuse. In particular, NSF can bind an energy source molecule, adenosine triphosphate (ATP), and the ATP-bound NSF develops internal tension via cleavage of ATP. This process is used to exert great force on SNARE complexes, eventually pulling them apart. However, although about 30 years have passed since the Nobel Prize winners’ discovery, how NSF/α-SNAP disassembled the SNARE complex remained a mystery to scientists due to a lack in methodology.
In a recent issue of Science, published on March 27, 2015, a research team, led by Tae-Young Yoon of the Department of Physics at the Korea Advanced Institute of Science and Technology (KAIST) and Reinhard Jahn of the Department of Neurobiology of the Max-Planck-Institute for Biophysical Chemistry, reports that NSF/α-SNAP disassemble a single SNARE complex using various single-molecule biophysical methods that allow them to monitor and manipulate individual protein complexes.
“We have learned that NSF releases energy in a burst within 20 milliseconds to “tear” the SNARE complex apart in a one-step global unfolding reaction, which is immediately followed by the release of SNARE proteins,” said Yoon.
Previously, it was believed that NSF disassembled a SNARE complex by unwinding it in a processive manner. Also, largely unexplained was how many cycles of ATP hydrolysis were required and how these cycles were connected to the disassembly of the SNARE complex.
Yoon added, “From our research, we found that NSF requires hydrolysis of ATPs that were already bound before it attached to the SNAREs—which means that only one round of an ATP turnover is sufficient for SNARE complex disassembly. Moreover, this is possible because NSF pulls a SNARE complex apart by building up the energy from individual ATPs and releasing it at once, yielding a “spring-loaded” mechanism.”
NSF is a member of the ATPases associated with various cellular activities family (AAA+ ATPase), which is essential for many cellular functions such as DNA replication and protein degradation, membrane fusion, microtubule severing, peroxisome biogenesis, signal transduction, and the regulation of gene expression. This research has added valuable new insights and hints for studying AAA+ ATPase proteins, which are crucial for various living beings.
The title of the research paper is “Spring-loaded unraveling of a single SNARE complex by NSF in one round of ATP turnover.” (DOI: 10.1126/science.aaa5267)
Youtube Link: https://www.youtube.com/watch?v=FqTSYHtyHWE&feature=youtu.be
Picture 1. Working model of how NSF/α-SNAP disassemble a single SNARE complex
Picture 2. After neurotransmitter release, NSF disassembles a single SNARE complex using a single round of ATP turnover in a single burst reaction.
2015.03.28 View 11848
Mystery in Membrane Traffic How NSF Disassembles Single SNAR Complex Solved
KAIST researchers discovered that the protein N-ethylmaleimide-sensitive factor (NSF) unravels a single SNARE complex using one round ATP turnover by tearing the complex with a single burst, contradicting a previous theory that it unwinds in a processive manner.
In 2013, James E. Rothman, Randy W. Schekman, and Thomas C. Südhof won the Nobel Prize in Physiology or Medicine for their discoveries of molecular machineries for vesicle trafficking, a major transport system in cells for maintaining cellular processes. Vesicle traffic acts as a kind of “home-delivery service” in cells. Vesicles package and deliver materials such as proteins and hormones from one cell organelle to another. Then it releases its contents by fusing with the target organelle’s membrane. One example of vesicle traffic is in neuronal communications, where neurotransmitters are released from a neuron. Some of the key proteins for vesicle traffic discovered by the Nobel Prize winners were N-ethylmaleimide-sensitive factor (NSF), alpha-soluble NSF attachment protein (α-SNAP), and soluble SNAP receptors (SNAREs).
SNARE proteins are known as the minimal machinery for membrane fusion. To induce membrane fusion, the proteins combine to form a SNARE complex in a four helical bundle, and NSF and α-SNAP disassemble the SNARE complex for reuse. In particular, NSF can bind an energy source molecule, adenosine triphosphate (ATP), and the ATP-bound NSF develops internal tension via cleavage of ATP. This process is used to exert great force on SNARE complexes, eventually pulling them apart. However, although about 30 years have passed since the Nobel Prize winners’ discovery, how NSF/α-SNAP disassembled the SNARE complex remained a mystery to scientists due to a lack in methodology.
In a recent issue of Science, published on March 27, 2015, a research team, led by Tae-Young Yoon of the Department of Physics at the Korea Advanced Institute of Science and Technology (KAIST) and Reinhard Jahn of the Department of Neurobiology of the Max-Planck-Institute for Biophysical Chemistry, reports that NSF/α-SNAP disassemble a single SNARE complex using various single-molecule biophysical methods that allow them to monitor and manipulate individual protein complexes.
“We have learned that NSF releases energy in a burst within 20 milliseconds to “tear” the SNARE complex apart in a one-step global unfolding reaction, which is immediately followed by the release of SNARE proteins,” said Yoon.
Previously, it was believed that NSF disassembled a SNARE complex by unwinding it in a processive manner. Also, largely unexplained was how many cycles of ATP hydrolysis were required and how these cycles were connected to the disassembly of the SNARE complex.
Yoon added, “From our research, we found that NSF requires hydrolysis of ATPs that were already bound before it attached to the SNAREs—which means that only one round of an ATP turnover is sufficient for SNARE complex disassembly. Moreover, this is possible because NSF pulls a SNARE complex apart by building up the energy from individual ATPs and releasing it at once, yielding a “spring-loaded” mechanism.”
NSF is a member of the ATPases associated with various cellular activities family (AAA+ ATPase), which is essential for many cellular functions such as DNA replication and protein degradation, membrane fusion, microtubule severing, peroxisome biogenesis, signal transduction, and the regulation of gene expression. This research has added valuable new insights and hints for studying AAA+ ATPase proteins, which are crucial for various living beings.
The title of the research paper is “Spring-loaded unraveling of a single SNARE complex by NSF in one round of ATP turnover.” (DOI: 10.1126/science.aaa5267)
Youtube Link: https://www.youtube.com/watch?v=FqTSYHtyHWE&feature=youtu.be
Picture 1. Working model of how NSF/α-SNAP disassemble a single SNARE complex
Picture 2. After neurotransmitter release, NSF disassembles a single SNARE complex using a single round of ATP turnover in a single burst reaction.
2015.03.28 View 11848 -
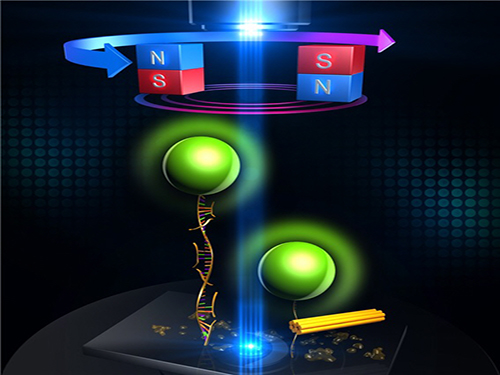 An Advanced Method of DNA Nanostructure Formation Developed
Professor Tae-Young Yoon’s research team from the Department of Physics at KAIST has developed a new method to form DNA nanostructures by using magnetic tweezers to observe and to induce the formation of the structure in real time.
Unlike traditional designs of "DNA origami" which relies on thermal or chemical annealing methods, the new technology utilizes a completely different dynamic in DNA folding. This allows the folding to be done within only ten minutes.
Developed in 2006, the "DNA origami" allows a long skeleton of DNA to be folded into an arbitrary structure by using small stapler DNA pieces. This has been a prominent method in DNA nanotechnology.
However, the traditional technology which adopts thermal processes could not control the DNA formation during the folding because every interaction among DNAs occurs simultaneously. Thus, the thermal processes, which take dozens of hours to complete, had to be repeated multiple times in order to find the optimal condition.
The research team designed a DNA folding using uni-molecular magnetic tweezers that applied force to a single DNA molecule while measuring the state of the DNA. Through this technology, they were able to induce the formation of DNA nanostructure and observe it at the same time.
During high temperature heat treatment, the first stage of conventional thermal processes, the internal structure of the long skeleton DNA untangles. To induce such state, after attaching one side of the skeleton DNA to the surface of glass and the other side to a magnetic material, the team unfolded the internal structure of the DNA by pulling the two sides apart with magnetic force.
Unlike the conventional thermal processes, this method lets the stapler DNA swiftly adhere to the skeleton DNA within a minute because the sites are revealed at room temperature.
After the stapler pieces connected to the skeleton, the team removed the magnetic force. Next, the structure folded through self-assembly as the stapler DNAs stuck to different sites on the skeleton DNA.
Professor Yoon said, “With the existing thermal methods, we could not differentiate the reactions of the DNA because the response of each DNA pieces mutually interacted with each other.” He added that “Using the magnetic tweezers, we were able to sort the process of DNA nanostructure formation into a series of reactions of DNA molecules that are well known, and shorten the time taken for formation in only ten minutes.”
He commented, “This nanostructure formation method will enable us to create more intricate and desirable DNA nanostructures by programming the folding of DNA origami structures.”
Conducted by Dr. Woori Bae under the guidance of Professor Yoon, the research findings were published online in the December 4th issue of Nature Communications.
Figure 1: Uni-molecular magnetic tweezers orchestrating the DNA nanostructure formation
Figure 2: The evolution of DNA nanostructure formation using magnetic tweezers. The DNA nanostructure with a 21-nanometer size was formed in about eight minutes.
2015.01.06 View 8056
An Advanced Method of DNA Nanostructure Formation Developed
Professor Tae-Young Yoon’s research team from the Department of Physics at KAIST has developed a new method to form DNA nanostructures by using magnetic tweezers to observe and to induce the formation of the structure in real time.
Unlike traditional designs of "DNA origami" which relies on thermal or chemical annealing methods, the new technology utilizes a completely different dynamic in DNA folding. This allows the folding to be done within only ten minutes.
Developed in 2006, the "DNA origami" allows a long skeleton of DNA to be folded into an arbitrary structure by using small stapler DNA pieces. This has been a prominent method in DNA nanotechnology.
However, the traditional technology which adopts thermal processes could not control the DNA formation during the folding because every interaction among DNAs occurs simultaneously. Thus, the thermal processes, which take dozens of hours to complete, had to be repeated multiple times in order to find the optimal condition.
The research team designed a DNA folding using uni-molecular magnetic tweezers that applied force to a single DNA molecule while measuring the state of the DNA. Through this technology, they were able to induce the formation of DNA nanostructure and observe it at the same time.
During high temperature heat treatment, the first stage of conventional thermal processes, the internal structure of the long skeleton DNA untangles. To induce such state, after attaching one side of the skeleton DNA to the surface of glass and the other side to a magnetic material, the team unfolded the internal structure of the DNA by pulling the two sides apart with magnetic force.
Unlike the conventional thermal processes, this method lets the stapler DNA swiftly adhere to the skeleton DNA within a minute because the sites are revealed at room temperature.
After the stapler pieces connected to the skeleton, the team removed the magnetic force. Next, the structure folded through self-assembly as the stapler DNAs stuck to different sites on the skeleton DNA.
Professor Yoon said, “With the existing thermal methods, we could not differentiate the reactions of the DNA because the response of each DNA pieces mutually interacted with each other.” He added that “Using the magnetic tweezers, we were able to sort the process of DNA nanostructure formation into a series of reactions of DNA molecules that are well known, and shorten the time taken for formation in only ten minutes.”
He commented, “This nanostructure formation method will enable us to create more intricate and desirable DNA nanostructures by programming the folding of DNA origami structures.”
Conducted by Dr. Woori Bae under the guidance of Professor Yoon, the research findings were published online in the December 4th issue of Nature Communications.
Figure 1: Uni-molecular magnetic tweezers orchestrating the DNA nanostructure formation
Figure 2: The evolution of DNA nanostructure formation using magnetic tweezers. The DNA nanostructure with a 21-nanometer size was formed in about eight minutes.
2015.01.06 View 8056