SCIE
-
 Professor Shin-Hyun Kim Receives the Young Scientist Award
Professor Shin-Hyun Kim from the Department of Chemical and Biomolecular Engineering received the Young Scientist Award from the Korean Academy of Science and Technology.
The Young Scientist Award is presented to a promising young Korean scientist under the age of 40 who shows significant potential, passion, and remarkable achievement.
Professor Kim was lauded for his research of intelligent soft materials. By applying his research, he developed a capsule sensor material that can not only be used for sensors, but also for displays, color aesthetics, anti-counterfeit technology, residual drug detection, and more.
The award ceremony took place on December 14 at the Gwacheon National Science Museum.
The Korean minister of Science and ICT delivered words of encouragement, reminding everyone that “the driving force behind creative performance of scientists is the provision of continuous support.” He added, “Researchers of Korea deserve greater public attention and support.”
(END)
2019.12.21 View 9351
Professor Shin-Hyun Kim Receives the Young Scientist Award
Professor Shin-Hyun Kim from the Department of Chemical and Biomolecular Engineering received the Young Scientist Award from the Korean Academy of Science and Technology.
The Young Scientist Award is presented to a promising young Korean scientist under the age of 40 who shows significant potential, passion, and remarkable achievement.
Professor Kim was lauded for his research of intelligent soft materials. By applying his research, he developed a capsule sensor material that can not only be used for sensors, but also for displays, color aesthetics, anti-counterfeit technology, residual drug detection, and more.
The award ceremony took place on December 14 at the Gwacheon National Science Museum.
The Korean minister of Science and ICT delivered words of encouragement, reminding everyone that “the driving force behind creative performance of scientists is the provision of continuous support.” He added, “Researchers of Korea deserve greater public attention and support.”
(END)
2019.12.21 View 9351 -
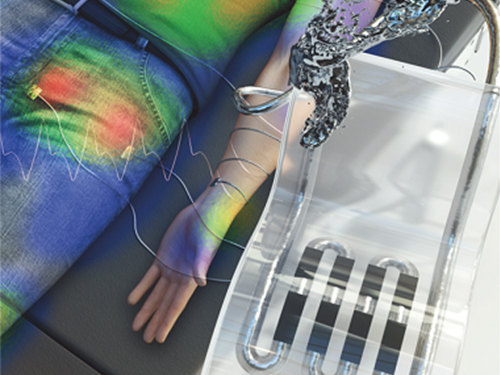 New Liquid Metal Wearable Pressure Sensor Created for Health Monitoring Applications
Soft pressure sensors have received significant research attention in a variety of fields, including soft robotics, electronic skin, and wearable electronics. Wearable soft pressure sensors have great potential for the real-time health monitoring and for the early diagnosis of diseases.
A KAIST research team led by Professor Inkyu Park from the Department of Mechanical Engineering developed a highly sensitive wearable pressure sensor for health monitoring applications. This work was reported in Advanced Healthcare Materials on November 21 as a front cover article.
This technology is capable of sensitive, precise, and continuous measurement of physiological and physical signals and shows great potential for health monitoring applications and the early diagnosis of diseases.
A soft pressure sensor is required to have high compliance, high sensitivity, low cost, long-term performance stability, and environmental stability in order to be employed for continuous health monitoring. Conventional solid-state soft pressure sensors using functional materials including carbon nanotubes and graphene have showed great sensing performance. However, these sensors suffer from limited stretchability, signal drifting, and long-term instability due to the distance between the stretchable substrate and the functional materials.
To overcome these issues, liquid-state electronics using liquid metal have been introduced for various wearable applications. Of these materials, Galinstan, a eutectic metal alloy of gallium, indium, and tin, has great mechanical and electrical properties that can be employed in wearable applications. But today’s liquid metal-based pressure sensors have low-pressure sensitivity, limiting their applicability for health monitoring devices.
The research team developed a 3D-printed rigid microbump array-integrated, liquid metal-based soft pressure sensor. With the help of 3D printing, the integration of a rigid microbump array and the master mold for a liquid metal microchannel could be achieved simultaneously, reducing the complexity of the manufacturing process. Through the integration of the rigid microbump and the microchannel, the new pressure sensor has an extremely low detection limit and enhanced pressure sensitivity compared to previously reported liquid metal-based pressure sensors. The proposed sensor also has a negligible signal drift over 10,000 cycles of pressure, bending, and stretching and exhibited excellent stability when subjected to various environmental conditions.
These performance outcomes make it an excellent sensor for various health monitoring devices. First, the research team demonstrated a wearable wristband device that can continuously monitor one’s pulse during exercise and be employed in a noninvasive cuffless BP monitoring system based on PTT calculations. Then, they introduced a wireless wearable heel pressure monitoring system that integrates three 3D-BLiPS with a wireless communication module.
Professor Park said, “It was possible to measure health indicators including pulse and blood pressure continuously as well as pressure of body parts using our proposed soft pressure sensor. We expect it to be used in health care applications, such as the prevention and the monitoring of the pressure-driven diseases such as pressure ulcers in the near future. There will be more opportunities for future research including a whole-body pressure monitoring system related to other physical parameters.”
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Ministry of Science and ICT.
< Figure 1. The front cover image of Advanced Healthcare Materials, Volume 8, Issue 22. >
< Figure 2. Highly sensitive liquid metal-based soft pressure sensor integrated with 3D-printed microbump array. >
< Figure 3. High pressure sensitivity and reliable sensing performances of the proposed sensor and wireless heel pressure monitoring application. >
-ProfileProfessor Inkyu ParkMicro/Nano Transducers Laboratoryhttp://mintlab1.kaist.ac.kr/
Department of Mechanical EngineeringKAIST
2019.12.20 View 16499
New Liquid Metal Wearable Pressure Sensor Created for Health Monitoring Applications
Soft pressure sensors have received significant research attention in a variety of fields, including soft robotics, electronic skin, and wearable electronics. Wearable soft pressure sensors have great potential for the real-time health monitoring and for the early diagnosis of diseases.
A KAIST research team led by Professor Inkyu Park from the Department of Mechanical Engineering developed a highly sensitive wearable pressure sensor for health monitoring applications. This work was reported in Advanced Healthcare Materials on November 21 as a front cover article.
This technology is capable of sensitive, precise, and continuous measurement of physiological and physical signals and shows great potential for health monitoring applications and the early diagnosis of diseases.
A soft pressure sensor is required to have high compliance, high sensitivity, low cost, long-term performance stability, and environmental stability in order to be employed for continuous health monitoring. Conventional solid-state soft pressure sensors using functional materials including carbon nanotubes and graphene have showed great sensing performance. However, these sensors suffer from limited stretchability, signal drifting, and long-term instability due to the distance between the stretchable substrate and the functional materials.
To overcome these issues, liquid-state electronics using liquid metal have been introduced for various wearable applications. Of these materials, Galinstan, a eutectic metal alloy of gallium, indium, and tin, has great mechanical and electrical properties that can be employed in wearable applications. But today’s liquid metal-based pressure sensors have low-pressure sensitivity, limiting their applicability for health monitoring devices.
The research team developed a 3D-printed rigid microbump array-integrated, liquid metal-based soft pressure sensor. With the help of 3D printing, the integration of a rigid microbump array and the master mold for a liquid metal microchannel could be achieved simultaneously, reducing the complexity of the manufacturing process. Through the integration of the rigid microbump and the microchannel, the new pressure sensor has an extremely low detection limit and enhanced pressure sensitivity compared to previously reported liquid metal-based pressure sensors. The proposed sensor also has a negligible signal drift over 10,000 cycles of pressure, bending, and stretching and exhibited excellent stability when subjected to various environmental conditions.
These performance outcomes make it an excellent sensor for various health monitoring devices. First, the research team demonstrated a wearable wristband device that can continuously monitor one’s pulse during exercise and be employed in a noninvasive cuffless BP monitoring system based on PTT calculations. Then, they introduced a wireless wearable heel pressure monitoring system that integrates three 3D-BLiPS with a wireless communication module.
Professor Park said, “It was possible to measure health indicators including pulse and blood pressure continuously as well as pressure of body parts using our proposed soft pressure sensor. We expect it to be used in health care applications, such as the prevention and the monitoring of the pressure-driven diseases such as pressure ulcers in the near future. There will be more opportunities for future research including a whole-body pressure monitoring system related to other physical parameters.”
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Ministry of Science and ICT.
< Figure 1. The front cover image of Advanced Healthcare Materials, Volume 8, Issue 22. >
< Figure 2. Highly sensitive liquid metal-based soft pressure sensor integrated with 3D-printed microbump array. >
< Figure 3. High pressure sensitivity and reliable sensing performances of the proposed sensor and wireless heel pressure monitoring application. >
-ProfileProfessor Inkyu ParkMicro/Nano Transducers Laboratoryhttp://mintlab1.kaist.ac.kr/
Department of Mechanical EngineeringKAIST
2019.12.20 View 16499 -
 Team Geumo Wins Consecutive Victories in K-Cyber Security Challenge
< Professor Sang Kil Cha >
< Masters Candidate Kangsu Kim and Researcher Corentin Soulet >
Team Geumo, led by Professor Sang Kil Cha from the Graduate School of Information Security, won the K-Cyber Security Challenge in the AI-based automatic vulnerability detection division for two consecutive years in 2018 and 2019.
The K-Cyber Security Challenge is an inter-machine hacking competition. Participants develop and operate AI-based systems that are capable of independently identifying software vulnerabilities and gaining operating rights through hacking. The K-Cyber Security Challenge, inspired by the US Cyber Grand Challenge launched by the Defense Advanced Research Projects Agency (DARPA), is hosted by the Ministry of Science and ICT and organized by the Korea Internet and Security Agency.
Researcher Corentin Soulet of the School of Computing and master’s student Kangsu Kim of the Graduate School of Information Security teamed up for the competition. Professor Cha, who has led the research on software and systems security since his days at Carnegie Mellon University, succeeded in establishing a world-class system using domestic technology.
In a recent collaboration with the Cyber Security Research Center, Professor Cha achieved a ten-fold increase in the speed of binary analysis engines, a key component of AI-based hacking systems. For this accomplishment, he received the Best Paper Award at the 2019 Network and Distributed System Security Workshop on Binary Analysis Research (NDSS BAR).
Kangsu Kim said, "It is a great honor to win the competition two years in a row. I will continue to work hard and apply my knowledge to serve society.”
(END)
2019.12.20 View 11676
Team Geumo Wins Consecutive Victories in K-Cyber Security Challenge
< Professor Sang Kil Cha >
< Masters Candidate Kangsu Kim and Researcher Corentin Soulet >
Team Geumo, led by Professor Sang Kil Cha from the Graduate School of Information Security, won the K-Cyber Security Challenge in the AI-based automatic vulnerability detection division for two consecutive years in 2018 and 2019.
The K-Cyber Security Challenge is an inter-machine hacking competition. Participants develop and operate AI-based systems that are capable of independently identifying software vulnerabilities and gaining operating rights through hacking. The K-Cyber Security Challenge, inspired by the US Cyber Grand Challenge launched by the Defense Advanced Research Projects Agency (DARPA), is hosted by the Ministry of Science and ICT and organized by the Korea Internet and Security Agency.
Researcher Corentin Soulet of the School of Computing and master’s student Kangsu Kim of the Graduate School of Information Security teamed up for the competition. Professor Cha, who has led the research on software and systems security since his days at Carnegie Mellon University, succeeded in establishing a world-class system using domestic technology.
In a recent collaboration with the Cyber Security Research Center, Professor Cha achieved a ten-fold increase in the speed of binary analysis engines, a key component of AI-based hacking systems. For this accomplishment, he received the Best Paper Award at the 2019 Network and Distributed System Security Workshop on Binary Analysis Research (NDSS BAR).
Kangsu Kim said, "It is a great honor to win the competition two years in a row. I will continue to work hard and apply my knowledge to serve society.”
(END)
2019.12.20 View 11676 -
 Professor Il-Doo Kim Named Scientist of the Year by the Journalists
Professor Il-Doo Kim from the Department of Materials Science and Engineering was named the 2019 Scientist of the Year by Korean science journalists. The award was conferred at the 2019 Science Press Night ceremony of the Korea Science Journalists Association (KSJA) on November 29.
Professor Kim focuses on developing nanofiber gas sensors for diagnosing diseases in advance by analyzing exhaled biomarkers with electrospinning technology. His outstanding research was praised and selected as one of the top 10 nanotechnology of 2019 by the Korea Nano Technology Research Society (KoNTRS), the Ministry of Science and ICT (MSIT), and the Ministry of Trade, Industry and Energy (MOTIE).
Professor Kim was honored with the QIAN Baojun Fiber Award, which is awarded every two years by Donghua University in Shanghai, China to recognize outstanding contributions in fiber science and technology. Professor Kim was also elected as an academician of the Asia Pacific Academy of Materials (APAM) on November 21 in Guangzhou, China.
In May, Professor Kim was appointed as an associate editor of ACS Nano, a leading international research journal in the field of nanoscience. In his editorial published in the May issue of ACS Nano, Professor Kim introduced and shared the history of KAIST and its vision for the future with other members of the journal. He hopes this will help with promoting a closer relationship between the members of the journal and KAIST moving forward.
“Above all,” he said in his acceptance speech, “the greatest news for me as an educator is that the first PhD graduate from our lab, Dr. Seonjin Choi, was appointed as the youngest professor in the Division of Materials Science and Engineering at Hanyang University on September 1.”
2019.12.17 View 13108
Professor Il-Doo Kim Named Scientist of the Year by the Journalists
Professor Il-Doo Kim from the Department of Materials Science and Engineering was named the 2019 Scientist of the Year by Korean science journalists. The award was conferred at the 2019 Science Press Night ceremony of the Korea Science Journalists Association (KSJA) on November 29.
Professor Kim focuses on developing nanofiber gas sensors for diagnosing diseases in advance by analyzing exhaled biomarkers with electrospinning technology. His outstanding research was praised and selected as one of the top 10 nanotechnology of 2019 by the Korea Nano Technology Research Society (KoNTRS), the Ministry of Science and ICT (MSIT), and the Ministry of Trade, Industry and Energy (MOTIE).
Professor Kim was honored with the QIAN Baojun Fiber Award, which is awarded every two years by Donghua University in Shanghai, China to recognize outstanding contributions in fiber science and technology. Professor Kim was also elected as an academician of the Asia Pacific Academy of Materials (APAM) on November 21 in Guangzhou, China.
In May, Professor Kim was appointed as an associate editor of ACS Nano, a leading international research journal in the field of nanoscience. In his editorial published in the May issue of ACS Nano, Professor Kim introduced and shared the history of KAIST and its vision for the future with other members of the journal. He hopes this will help with promoting a closer relationship between the members of the journal and KAIST moving forward.
“Above all,” he said in his acceptance speech, “the greatest news for me as an educator is that the first PhD graduate from our lab, Dr. Seonjin Choi, was appointed as the youngest professor in the Division of Materials Science and Engineering at Hanyang University on September 1.”
2019.12.17 View 13108 -
 New Members of KAST 2020
< Professor Zong-Tae Bae (Left) and Professor Sang Ouk Kim (Right) >
Professor Zong-Tae Bae from the School of Management Engineering and Professor Sang Ouk Kim from the Department of Materials Science and Engineering became new fellows of the Korean Academy of Science and Technology (KAST) along with 22 other scientists in Korea.
On November 22, KAST announced 24 new members for the year 2020. This includes seven scientists from the field of natural sciences, six from engineering, four from medical sciences, another four from policy research, and three from agriculture and fishery.
The new fellows will begin their term from January next year, and their fellowships wll be conferred during the KAST’s New Year Reception to be held on January 14 in Seoul.
(END)
2019.12.09 View 14623
New Members of KAST 2020
< Professor Zong-Tae Bae (Left) and Professor Sang Ouk Kim (Right) >
Professor Zong-Tae Bae from the School of Management Engineering and Professor Sang Ouk Kim from the Department of Materials Science and Engineering became new fellows of the Korean Academy of Science and Technology (KAST) along with 22 other scientists in Korea.
On November 22, KAST announced 24 new members for the year 2020. This includes seven scientists from the field of natural sciences, six from engineering, four from medical sciences, another four from policy research, and three from agriculture and fishery.
The new fellows will begin their term from January next year, and their fellowships wll be conferred during the KAST’s New Year Reception to be held on January 14 in Seoul.
(END)
2019.12.09 View 14623 -
 AI to Determine When to Intervene with Your Driving
(Professor Uichin Lee (left) and PhD candidate Auk Kim)
Can your AI agent judge when to talk to you while you are driving? According to a KAIST research team, their in-vehicle conservation service technology will judge when it is appropriate to contact you to ensure your safety.
Professor Uichin Lee from the Department of Industrial and Systems Engineering at KAIST and his research team have developed AI technology that automatically detects safe moments for AI agents to provide conversation services to drivers.
Their research focuses on solving the potential problems of distraction created by in-vehicle conversation services. If an AI agent talks to a driver at an inopportune moment, such as while making a turn, a car accident will be more likely to occur.
In-vehicle conversation services need to be convenient as well as safe. However, the cognitive burden of multitasking negatively influences the quality of the service. Users tend to be more distracted during certain traffic conditions. To address this long-standing challenge of the in-vehicle conversation services, the team introduced a composite cognitive model that considers both safe driving and auditory-verbal service performance and used a machine-learning model for all collected data.
The combination of these individual measures is able to determine the appropriate moments for conversation and most appropriate types of conversational services. For instance, in the case of delivering simple-context information, such as a weather forecast, driver safety alone would be the most appropriate consideration. Meanwhile, when delivering information that requires a driver response, such as a “Yes” or “No,” the combination of driver safety and auditory-verbal performance should be considered.
The research team developed a prototype of an in-vehicle conversation service based on a navigation app that can be used in real driving environments. The app was also connected to the vehicle to collect in-vehicle OBD-II/CAN data, such as the steering wheel angle and brake pedal position, and mobility and environmental data such as the distance between successive cars and traffic flow.
Using pseudo-conversation services, the research team collected a real-world driving dataset consisting of 1,388 interactions and sensor data from 29 drivers who interacted with AI conversational agents. Machine learning analysis based on the dataset demonstrated that the opportune moments for driver interruption could be correctly inferred with 87% accuracy.
The safety enhancement technology developed by the team is expected to minimize driver distractions caused by in-vehicle conversation services. This technology can be directly applied to current in-vehicle systems that provide conversation services. It can also be extended and applied to the real-time detection of driver distraction problems caused by the use of a smartphone while driving.
Professor Lee said, “In the near future, cars will proactively deliver various in-vehicle conversation services. This technology will certainly help vehicles interact with their drivers safely as it can fairly accurately determine when to provide conversation services using only basic sensor data generated by cars.”
The researchers presented their findings at the ACM International Joint Conference on Pervasive and Ubiquitous Computing (Ubicomp’19) in London, UK. This research was supported in part by Hyundai NGV and by the Next-Generation Information Computing Development Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT.
(Figure: Visual description of safe enhancement technology for in-vehicle conversation services)
2019.11.13 View 19889
AI to Determine When to Intervene with Your Driving
(Professor Uichin Lee (left) and PhD candidate Auk Kim)
Can your AI agent judge when to talk to you while you are driving? According to a KAIST research team, their in-vehicle conservation service technology will judge when it is appropriate to contact you to ensure your safety.
Professor Uichin Lee from the Department of Industrial and Systems Engineering at KAIST and his research team have developed AI technology that automatically detects safe moments for AI agents to provide conversation services to drivers.
Their research focuses on solving the potential problems of distraction created by in-vehicle conversation services. If an AI agent talks to a driver at an inopportune moment, such as while making a turn, a car accident will be more likely to occur.
In-vehicle conversation services need to be convenient as well as safe. However, the cognitive burden of multitasking negatively influences the quality of the service. Users tend to be more distracted during certain traffic conditions. To address this long-standing challenge of the in-vehicle conversation services, the team introduced a composite cognitive model that considers both safe driving and auditory-verbal service performance and used a machine-learning model for all collected data.
The combination of these individual measures is able to determine the appropriate moments for conversation and most appropriate types of conversational services. For instance, in the case of delivering simple-context information, such as a weather forecast, driver safety alone would be the most appropriate consideration. Meanwhile, when delivering information that requires a driver response, such as a “Yes” or “No,” the combination of driver safety and auditory-verbal performance should be considered.
The research team developed a prototype of an in-vehicle conversation service based on a navigation app that can be used in real driving environments. The app was also connected to the vehicle to collect in-vehicle OBD-II/CAN data, such as the steering wheel angle and brake pedal position, and mobility and environmental data such as the distance between successive cars and traffic flow.
Using pseudo-conversation services, the research team collected a real-world driving dataset consisting of 1,388 interactions and sensor data from 29 drivers who interacted with AI conversational agents. Machine learning analysis based on the dataset demonstrated that the opportune moments for driver interruption could be correctly inferred with 87% accuracy.
The safety enhancement technology developed by the team is expected to minimize driver distractions caused by in-vehicle conversation services. This technology can be directly applied to current in-vehicle systems that provide conversation services. It can also be extended and applied to the real-time detection of driver distraction problems caused by the use of a smartphone while driving.
Professor Lee said, “In the near future, cars will proactively deliver various in-vehicle conversation services. This technology will certainly help vehicles interact with their drivers safely as it can fairly accurately determine when to provide conversation services using only basic sensor data generated by cars.”
The researchers presented their findings at the ACM International Joint Conference on Pervasive and Ubiquitous Computing (Ubicomp’19) in London, UK. This research was supported in part by Hyundai NGV and by the Next-Generation Information Computing Development Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT.
(Figure: Visual description of safe enhancement technology for in-vehicle conversation services)
2019.11.13 View 19889 -
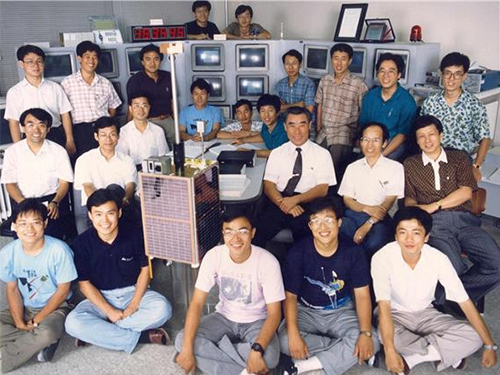 SaTReC, Birthplace of Korea’s First Satellite, Celebrates 30th Anniversary
< SaTReC researchers who developed Korea's first satellite, KITSAT-1 >
The Satellite Technology Research Center (SaTReC) at KAIST, which launched the Korea’s first satellite KITSAT-1, celebrated 30 years in operation last week. A ceremony in honor of this milestone was held on campus on October 30. With the launching of KITSAT-1 in 1992, SaTReC paved the way for space research in Korea, and helped the nation achieve technological independence and strengthen competitiveness in the field.
The ceremony was attended by over 100 affiliates from academia and industry, including the family of the late Dr. Soon-dal Choi, the first director of SaTReC also known as the father of the first Korean satellite KITSAT-1 (nicknamed “Our Star” in Korean). His family members traveled all the way from the US to Korea for the event. A plaque of appreciation was posthumously awarded to the family of former Director Choi in memory of his pioneering Korean satellite research.
Right after the establishment of SaTReC in 1989, Dr.Choi dispatched five KAIST students to the University of Surrey in the UK to develop the Korea’s first satellite KITSAT-1 under a bilateral agreement for a joint research program.
KITSAT-1, completed in collaboration with Surrey researchers, was successfully launched from the Guiana Space Center in August 1992. Through this launch, Korea became the 22nd nation to own a satellite, and launched the domestically produced follow-up satellite KITSAT-2 in September 1993.
Since then, SaTReC has developed a total of nine satellites, including three in the KITSAT series in the 1990s as well as five STSATs and one Next-Generation Small Satellite in the 2000s. These satellites are still in operation today, thanks to SaTReC’s constant maintenance.
SaTReC is still contributing to the verification of core space technologies and Earth and space observation technologies using small satellites. It is also training specialized personnel in national space research and development.
Most significantly, STSAT-2C, also commonly known as the Naro Science Satellite, was launched on January 30, 2013 and served an important role in allowing the first Korean launch vehicle Naro-1 (KSLV-1) to enter into orbit.
SaTReC researchers are now working on developing a Next-Generation Small Satellite named NEXTSat-2 that boasts a Synthetic Aperture Radar (SAR) system developed with domestic technology. NEXTSat-2 will be launched in 2022 from Korean soil, carried by a Korean launch vehicle developed with local technology.
Director of SaTReC Sejin Kwon said, “We will follow the noble spirit of the late Dr. Soon-dal Choi, who dedicated his entire life to the nation’s satellite research and bolstered our commitment to the development of Korea’s future space technology.” He added, “We will pursue our dreams of space exploration with a sense of social responsibility to pay back to society the benefits reaped from space technology.”
The ceremony was followed by a Future Space Technology Workshop, where eight KAIST professors participated as speakers.
< Timeline of Korea's Satellite Research and Development >
(END)
2019.11.05 View 7757
SaTReC, Birthplace of Korea’s First Satellite, Celebrates 30th Anniversary
< SaTReC researchers who developed Korea's first satellite, KITSAT-1 >
The Satellite Technology Research Center (SaTReC) at KAIST, which launched the Korea’s first satellite KITSAT-1, celebrated 30 years in operation last week. A ceremony in honor of this milestone was held on campus on October 30. With the launching of KITSAT-1 in 1992, SaTReC paved the way for space research in Korea, and helped the nation achieve technological independence and strengthen competitiveness in the field.
The ceremony was attended by over 100 affiliates from academia and industry, including the family of the late Dr. Soon-dal Choi, the first director of SaTReC also known as the father of the first Korean satellite KITSAT-1 (nicknamed “Our Star” in Korean). His family members traveled all the way from the US to Korea for the event. A plaque of appreciation was posthumously awarded to the family of former Director Choi in memory of his pioneering Korean satellite research.
Right after the establishment of SaTReC in 1989, Dr.Choi dispatched five KAIST students to the University of Surrey in the UK to develop the Korea’s first satellite KITSAT-1 under a bilateral agreement for a joint research program.
KITSAT-1, completed in collaboration with Surrey researchers, was successfully launched from the Guiana Space Center in August 1992. Through this launch, Korea became the 22nd nation to own a satellite, and launched the domestically produced follow-up satellite KITSAT-2 in September 1993.
Since then, SaTReC has developed a total of nine satellites, including three in the KITSAT series in the 1990s as well as five STSATs and one Next-Generation Small Satellite in the 2000s. These satellites are still in operation today, thanks to SaTReC’s constant maintenance.
SaTReC is still contributing to the verification of core space technologies and Earth and space observation technologies using small satellites. It is also training specialized personnel in national space research and development.
Most significantly, STSAT-2C, also commonly known as the Naro Science Satellite, was launched on January 30, 2013 and served an important role in allowing the first Korean launch vehicle Naro-1 (KSLV-1) to enter into orbit.
SaTReC researchers are now working on developing a Next-Generation Small Satellite named NEXTSat-2 that boasts a Synthetic Aperture Radar (SAR) system developed with domestic technology. NEXTSat-2 will be launched in 2022 from Korean soil, carried by a Korean launch vehicle developed with local technology.
Director of SaTReC Sejin Kwon said, “We will follow the noble spirit of the late Dr. Soon-dal Choi, who dedicated his entire life to the nation’s satellite research and bolstered our commitment to the development of Korea’s future space technology.” He added, “We will pursue our dreams of space exploration with a sense of social responsibility to pay back to society the benefits reaped from space technology.”
The ceremony was followed by a Future Space Technology Workshop, where eight KAIST professors participated as speakers.
< Timeline of Korea's Satellite Research and Development >
(END)
2019.11.05 View 7757 -
 Tungsten Suboxide Improves the Efficiency of Platinum in Hydrogen Production
< PhD Candidate Jinkyu Park and Professor Jinwoo Lee >
Researchers presented a new strategy for enhancing catalytic activity using tungsten suboxide as a single-atom catalyst (SAC). This strategy, which significantly improves hydrogen evolution reaction (HER) in metal platinum (pt) by 16.3 times, sheds light on the development of new electrochemical catalyst technologies.
Hydrogen has been touted as a promising alternative to fossil fuels. However, most of the conventional industrial hydrogen production methods come with environmental issues, releasing significant amounts of carbon dioxide and greenhouse gases.
Electrochemical water splitting is considered a potential approach for clean hydrogen production. Pt is one of the most commonly used catalysts to improve HER performance in electrochemical water splitting, but the high cost and scarcity of Pt remain key obstacles to mass commercial applications.
SACs, where all metal species are individually dispersed on a desired support material, have been identified as one way to reduce the amount of Pt usage, as they offer the maximum number of surface exposed Pt atoms.
Inspired by earlier studies, which mainly focused on SACs supported by carbon-based materials, a KAIST research team led by Professor Jinwoo Lee from the Department of Chemical and Biomolecular Engineering investigated the influence of support materials on the performance of SACs.
Professor Lee and his researchers suggested mesoporous tungsten suboxide as a new support material for atomically dispersed Pt, as this was expected to provide high electronic conductivity and have a synergetic effect with Pt.
They compared the performance of single-atom Pt supported by carbon and tungsten suboxide respectively. The results revealed that the support effect occurred with tungsten suboxide, in which the mass activity of a single-atom Pt supported by tungsten suboxide was 2.1 times greater than that of single-atom Pt supported by carbon, and 16.3 times higher than that of Pt nanoparticles supported by carbon.
The team indicated a change in the electronic structure of Pt via charge transfer from tungsten suboxide to Pt. This phenomenon was reported as a result of strong metal-support interaction between Pt and tungsten suboxide.
HER performance can be improved not only by changing the electronic structure of the supported metal, but also by inducing another support effect, the spillover effect, the research group reported. Hydrogen spillover is a phenomenon where adsorbed hydrogen migrates from one surface to another, and it occurs more easily as the Pt size becomes smaller.
The researchers compared the performance of single-atom Pt and Pt nanoparticles supported by tungsten suboxide. The single-atom Pt supported by tungsten suboxide exhibited a higher degree of hydrogen spillover phenomenon, which enhanced the Pt mass activity for hydrogen evolution up to 10.7 times compared to Pt nanoparticles supported by tungsten suboxide.
Professor Lee said, “Choosing the right support material is important for improving electrocatalysis in hydrogen production. The tungsten suboxide catalyst we used to support Pt in our study implies that interactions between the well-matched metal and support can drastically enhance the efficiency of the process.”
This research was supported by the Ministry of Science and ICT and introduced in the International Edition of the German journal Angewandte Chemie.
Figure. Schematic representation of hydrogen evolution reaction (HER) of pseudo single-atom Pt supported by tungsten suboxide
-Publication
Jinkyu Park, Dr. Seonggyu Lee, Hee-Eun Kim, Ara Cho, Seongbeen Kim, Dr. Youngjin Ye, Prof. Jeong Woo Han, Prof. Hyunjoo Lee, Dr. Jong Hyun Jang, and Prof. Jinwoo Lee. 2019. Investigation of the Support Effect in Atomically Dispersed Pt on WO3−x for Utilization of Pt in the Hydrogen Evolution Reaction. International Edition of Angewandte Chemie. Volume No. 58. Issue No. 45. 6 pages. https://doi.org/10.1002/anie.201908122
-ProfileProfessor Jinwoo LeeConvergence of Energy and Nano Science Laboratoryhttp://cens.kaist.ac.kr
Department of Chemical and Biomolecular EngineeringKAIST
2019.10.28 View 23213
Tungsten Suboxide Improves the Efficiency of Platinum in Hydrogen Production
< PhD Candidate Jinkyu Park and Professor Jinwoo Lee >
Researchers presented a new strategy for enhancing catalytic activity using tungsten suboxide as a single-atom catalyst (SAC). This strategy, which significantly improves hydrogen evolution reaction (HER) in metal platinum (pt) by 16.3 times, sheds light on the development of new electrochemical catalyst technologies.
Hydrogen has been touted as a promising alternative to fossil fuels. However, most of the conventional industrial hydrogen production methods come with environmental issues, releasing significant amounts of carbon dioxide and greenhouse gases.
Electrochemical water splitting is considered a potential approach for clean hydrogen production. Pt is one of the most commonly used catalysts to improve HER performance in electrochemical water splitting, but the high cost and scarcity of Pt remain key obstacles to mass commercial applications.
SACs, where all metal species are individually dispersed on a desired support material, have been identified as one way to reduce the amount of Pt usage, as they offer the maximum number of surface exposed Pt atoms.
Inspired by earlier studies, which mainly focused on SACs supported by carbon-based materials, a KAIST research team led by Professor Jinwoo Lee from the Department of Chemical and Biomolecular Engineering investigated the influence of support materials on the performance of SACs.
Professor Lee and his researchers suggested mesoporous tungsten suboxide as a new support material for atomically dispersed Pt, as this was expected to provide high electronic conductivity and have a synergetic effect with Pt.
They compared the performance of single-atom Pt supported by carbon and tungsten suboxide respectively. The results revealed that the support effect occurred with tungsten suboxide, in which the mass activity of a single-atom Pt supported by tungsten suboxide was 2.1 times greater than that of single-atom Pt supported by carbon, and 16.3 times higher than that of Pt nanoparticles supported by carbon.
The team indicated a change in the electronic structure of Pt via charge transfer from tungsten suboxide to Pt. This phenomenon was reported as a result of strong metal-support interaction between Pt and tungsten suboxide.
HER performance can be improved not only by changing the electronic structure of the supported metal, but also by inducing another support effect, the spillover effect, the research group reported. Hydrogen spillover is a phenomenon where adsorbed hydrogen migrates from one surface to another, and it occurs more easily as the Pt size becomes smaller.
The researchers compared the performance of single-atom Pt and Pt nanoparticles supported by tungsten suboxide. The single-atom Pt supported by tungsten suboxide exhibited a higher degree of hydrogen spillover phenomenon, which enhanced the Pt mass activity for hydrogen evolution up to 10.7 times compared to Pt nanoparticles supported by tungsten suboxide.
Professor Lee said, “Choosing the right support material is important for improving electrocatalysis in hydrogen production. The tungsten suboxide catalyst we used to support Pt in our study implies that interactions between the well-matched metal and support can drastically enhance the efficiency of the process.”
This research was supported by the Ministry of Science and ICT and introduced in the International Edition of the German journal Angewandte Chemie.
Figure. Schematic representation of hydrogen evolution reaction (HER) of pseudo single-atom Pt supported by tungsten suboxide
-Publication
Jinkyu Park, Dr. Seonggyu Lee, Hee-Eun Kim, Ara Cho, Seongbeen Kim, Dr. Youngjin Ye, Prof. Jeong Woo Han, Prof. Hyunjoo Lee, Dr. Jong Hyun Jang, and Prof. Jinwoo Lee. 2019. Investigation of the Support Effect in Atomically Dispersed Pt on WO3−x for Utilization of Pt in the Hydrogen Evolution Reaction. International Edition of Angewandte Chemie. Volume No. 58. Issue No. 45. 6 pages. https://doi.org/10.1002/anie.201908122
-ProfileProfessor Jinwoo LeeConvergence of Energy and Nano Science Laboratoryhttp://cens.kaist.ac.kr
Department of Chemical and Biomolecular EngineeringKAIST
2019.10.28 View 23213 -
 Image Analysis to Automatically Quantify Gender Bias in Movies
Many commercial films worldwide continue to express womanhood in a stereotypical manner, a recent study using image analysis showed. A KAIST research team developed a novel image analysis method for automatically quantifying the degree of gender bias in films.
The ‘Bechdel Test’ has been the most representative and general method of evaluating gender bias in films. This test indicates the degree of gender bias in a film by measuring how active the presence of women is in a film. A film passes the Bechdel Test if the film (1) has at least two female characters, (2) who talk to each other, and (3) their conversation is not related to the male characters.
However, the Bechdel Test has fundamental limitations regarding the accuracy and practicality of the evaluation. Firstly, the Bechdel Test requires considerable human resources, as it is performed subjectively by a person. More importantly, the Bechdel Test analyzes only a single aspect of the film, the dialogues between characters in the script, and provides only a dichotomous result of passing the test, neglecting the fact that a film is a visual art form reflecting multi-layered and complicated gender bias phenomena. It is also difficult to fully represent today’s various discourse on gender bias, which is much more diverse than in 1985 when the Bechdel Test was first presented.
Inspired by these limitations, a KAIST research team led by Professor Byungjoo Lee from the Graduate School of Culture Technology proposed an advanced system that uses computer vision technology to automatically analyzes the visual information of each frame of the film. This allows the system to more accurately and practically evaluate the degree to which female and male characters are discriminatingly depicted in a film in quantitative terms, and further enables the revealing of gender bias that conventional analysis methods could not yet detect.
Professor Lee and his researchers Ji Yoon Jang and Sangyoon Lee analyzed 40 films from Hollywood and South Korea released between 2017 and 2018. They downsampled the films from 24 to 3 frames per second, and used Microsoft’s Face API facial recognition technology and object detection technology YOLO9000 to verify the details of the characters and their surrounding objects in the scenes.
Using the new system, the team computed eight quantitative indices that describe the representation of a particular gender in the films. They are: emotional diversity, spatial staticity, spatial occupancy, temporal occupancy, mean age, intellectual image, emphasis on appearance, and type and frequency of surrounding objects.
Figure 1. System Diagram
Figure 2. 40 Hollywood and Korean Films Analyzed in the Study
According to the emotional diversity index, the depicted women were found to be more prone to expressing passive emotions, such as sadness, fear, and surprise. In contrast, male characters in the same films were more likely to demonstrate active emotions, such as anger and hatred.
Figure 3. Difference in Emotional Diversity between Female and Male Characters
The type and frequency of surrounding objects index revealed that female characters and automobiles were tracked together only 55.7 % as much as that of male characters, while they were more likely to appear with furniture and in a household, with 123.9% probability.
In cases of temporal occupancy and mean age, female characters appeared less frequently in films than males at the rate of 56%, and were on average younger in 79.1% of the cases. These two indices were especially conspicuous in Korean films.
Professor Lee said, “Our research confirmed that many commercial films depict women from a stereotypical perspective. I hope this result promotes public awareness of the importance of taking prudence when filmmakers create characters in films.”
This study was supported by KAIST College of Liberal Arts and Convergence Science as part of the Venture Research Program for Master’s and PhD Students, and will be presented at the 22nd ACM Conference on Computer-Supported Cooperative Work and Social Computing (CSCW) on November 11 to be held in Austin, Texas.
Publication:
Ji Yoon Jang, Sangyoon Lee, and Byungjoo Lee. 2019. Quantification of Gender Representation Bias in Commercial Films based on Image Analysis. In Proceedings of the 22nd ACM Conference on Computer-Supported Cooperative Work and Social Computing (CSCW). ACM, New York, NY, USA, Article 198, 29 pages. https://doi.org/10.1145/3359300
Link to download the full-text paper:
https://files.cargocollective.com/611692/cscw198-jangA--1-.pdf
Profile: Prof. Byungjoo Lee, MD, PhD
byungjoo.lee@kaist.ac.kr
http://kiml.org/
Assistant Professor
Graduate School of Culture Technology (CT)
Korea Advanced Institute of Science and Technology (KAIST)
https://www.kaist.ac.kr Daejeon 34141, Korea
Profile: Ji Yoon Jang, M.S.
yoone3422@kaist.ac.kr
Interactive Media Lab
Graduate School of Culture Technology (CT)
Korea Advanced Institute of Science and Technology (KAIST)
https://www.kaist.ac.kr Daejeon 34141, Korea
Profile: Sangyoon Lee, M.S. Candidate
sl2820@kaist.ac.kr
Interactive Media Lab
Graduate School of Culture Technology (CT)
Korea Advanced Institute of Science and Technology (KAIST)
https://www.kaist.ac.kr Daejeon 34141, Korea
(END)
2019.10.17 View 28085
Image Analysis to Automatically Quantify Gender Bias in Movies
Many commercial films worldwide continue to express womanhood in a stereotypical manner, a recent study using image analysis showed. A KAIST research team developed a novel image analysis method for automatically quantifying the degree of gender bias in films.
The ‘Bechdel Test’ has been the most representative and general method of evaluating gender bias in films. This test indicates the degree of gender bias in a film by measuring how active the presence of women is in a film. A film passes the Bechdel Test if the film (1) has at least two female characters, (2) who talk to each other, and (3) their conversation is not related to the male characters.
However, the Bechdel Test has fundamental limitations regarding the accuracy and practicality of the evaluation. Firstly, the Bechdel Test requires considerable human resources, as it is performed subjectively by a person. More importantly, the Bechdel Test analyzes only a single aspect of the film, the dialogues between characters in the script, and provides only a dichotomous result of passing the test, neglecting the fact that a film is a visual art form reflecting multi-layered and complicated gender bias phenomena. It is also difficult to fully represent today’s various discourse on gender bias, which is much more diverse than in 1985 when the Bechdel Test was first presented.
Inspired by these limitations, a KAIST research team led by Professor Byungjoo Lee from the Graduate School of Culture Technology proposed an advanced system that uses computer vision technology to automatically analyzes the visual information of each frame of the film. This allows the system to more accurately and practically evaluate the degree to which female and male characters are discriminatingly depicted in a film in quantitative terms, and further enables the revealing of gender bias that conventional analysis methods could not yet detect.
Professor Lee and his researchers Ji Yoon Jang and Sangyoon Lee analyzed 40 films from Hollywood and South Korea released between 2017 and 2018. They downsampled the films from 24 to 3 frames per second, and used Microsoft’s Face API facial recognition technology and object detection technology YOLO9000 to verify the details of the characters and their surrounding objects in the scenes.
Using the new system, the team computed eight quantitative indices that describe the representation of a particular gender in the films. They are: emotional diversity, spatial staticity, spatial occupancy, temporal occupancy, mean age, intellectual image, emphasis on appearance, and type and frequency of surrounding objects.
Figure 1. System Diagram
Figure 2. 40 Hollywood and Korean Films Analyzed in the Study
According to the emotional diversity index, the depicted women were found to be more prone to expressing passive emotions, such as sadness, fear, and surprise. In contrast, male characters in the same films were more likely to demonstrate active emotions, such as anger and hatred.
Figure 3. Difference in Emotional Diversity between Female and Male Characters
The type and frequency of surrounding objects index revealed that female characters and automobiles were tracked together only 55.7 % as much as that of male characters, while they were more likely to appear with furniture and in a household, with 123.9% probability.
In cases of temporal occupancy and mean age, female characters appeared less frequently in films than males at the rate of 56%, and were on average younger in 79.1% of the cases. These two indices were especially conspicuous in Korean films.
Professor Lee said, “Our research confirmed that many commercial films depict women from a stereotypical perspective. I hope this result promotes public awareness of the importance of taking prudence when filmmakers create characters in films.”
This study was supported by KAIST College of Liberal Arts and Convergence Science as part of the Venture Research Program for Master’s and PhD Students, and will be presented at the 22nd ACM Conference on Computer-Supported Cooperative Work and Social Computing (CSCW) on November 11 to be held in Austin, Texas.
Publication:
Ji Yoon Jang, Sangyoon Lee, and Byungjoo Lee. 2019. Quantification of Gender Representation Bias in Commercial Films based on Image Analysis. In Proceedings of the 22nd ACM Conference on Computer-Supported Cooperative Work and Social Computing (CSCW). ACM, New York, NY, USA, Article 198, 29 pages. https://doi.org/10.1145/3359300
Link to download the full-text paper:
https://files.cargocollective.com/611692/cscw198-jangA--1-.pdf
Profile: Prof. Byungjoo Lee, MD, PhD
byungjoo.lee@kaist.ac.kr
http://kiml.org/
Assistant Professor
Graduate School of Culture Technology (CT)
Korea Advanced Institute of Science and Technology (KAIST)
https://www.kaist.ac.kr Daejeon 34141, Korea
Profile: Ji Yoon Jang, M.S.
yoone3422@kaist.ac.kr
Interactive Media Lab
Graduate School of Culture Technology (CT)
Korea Advanced Institute of Science and Technology (KAIST)
https://www.kaist.ac.kr Daejeon 34141, Korea
Profile: Sangyoon Lee, M.S. Candidate
sl2820@kaist.ac.kr
Interactive Media Lab
Graduate School of Culture Technology (CT)
Korea Advanced Institute of Science and Technology (KAIST)
https://www.kaist.ac.kr Daejeon 34141, Korea
(END)
2019.10.17 View 28085 -
 A Mathematical Model Reveals Long-Distance Cell Communication Mechanism
How can tens of thousands of people in a large football stadium all clap together with the same beat even though they can only hear the people near them clapping?
A combination of a partial differential equation and a synthetic circuit in microbes answers this question. An interdisciplinary collaborative team of Professor Jae Kyoung Kim at KAIST, Professor Krešimir Josić at the University of Houston, and Professor Matt Bennett at Rice University has identified how a large community can communicate with each other almost simultaneously even with very short distance signaling. The research was reported at Nature Chemical Biology.
Cells often communicate using signaling molecules, which can travel only a short distance. Nevertheless, the cells can also communicate over large distances to spur collective action. The team revealed a cell communication mechanism that quickly forms a network of local interactions to spur collective action, even in large communities.
The research team used an engineered transcriptional circuit of combined positive and negative feedback loops in E. coli, which can periodically release two types of signaling molecules: activator and repressor. As the signaling molecules travel over a short distance, cells can only talk to their nearest neighbors. However, cell communities synchronize oscillatory gene expression in spatially extended systems as long as the transcriptional circuit contains a positive feedback loop for the activator.
Professor Kim said that analyzing and understanding such high-dimensional dynamics was extremely difficult. He explained, “That’s why we used high-dimensional partial differential equation to describe the system based on the interactions among various types of molecules.” Surprisingly, the mathematical model accurately simulates the synthesis of the signaling molecules in the cell and their spatial diffusion throughout the chamber and their effect on neighboring cells.
The team simplified the high-dimensional system into a one-dimensional orbit, noting that the system repeats periodically. This allowed them to discover that cells can make one voice when they lowered their own voice and listened to the others. “It turns out the positive feedback loop reduces the distance between moving points and finally makes them move all together. That’s why you clap louder when you hear applause from nearby neighbors and everyone eventually claps together at almost the same time,” said Professor Kim.
Professor Kim added, “Math is a powerful as it simplifies complex thing so that we can find an essential underlying property. This finding would not have been possible without the simplification of complex systems using mathematics."
The National Institutes of Health, the National Science Foundation, the Robert A. Welch Foundation, the Hamill Foundation, the National Research Foundation of Korea, and the T.J. Park Science Fellowship of POSCO supported the research.
(Figure: Complex molecular interactions among microbial consortia is simplified as interactions among points on a limit cycle (right).)
2019.10.15 View 29979
A Mathematical Model Reveals Long-Distance Cell Communication Mechanism
How can tens of thousands of people in a large football stadium all clap together with the same beat even though they can only hear the people near them clapping?
A combination of a partial differential equation and a synthetic circuit in microbes answers this question. An interdisciplinary collaborative team of Professor Jae Kyoung Kim at KAIST, Professor Krešimir Josić at the University of Houston, and Professor Matt Bennett at Rice University has identified how a large community can communicate with each other almost simultaneously even with very short distance signaling. The research was reported at Nature Chemical Biology.
Cells often communicate using signaling molecules, which can travel only a short distance. Nevertheless, the cells can also communicate over large distances to spur collective action. The team revealed a cell communication mechanism that quickly forms a network of local interactions to spur collective action, even in large communities.
The research team used an engineered transcriptional circuit of combined positive and negative feedback loops in E. coli, which can periodically release two types of signaling molecules: activator and repressor. As the signaling molecules travel over a short distance, cells can only talk to their nearest neighbors. However, cell communities synchronize oscillatory gene expression in spatially extended systems as long as the transcriptional circuit contains a positive feedback loop for the activator.
Professor Kim said that analyzing and understanding such high-dimensional dynamics was extremely difficult. He explained, “That’s why we used high-dimensional partial differential equation to describe the system based on the interactions among various types of molecules.” Surprisingly, the mathematical model accurately simulates the synthesis of the signaling molecules in the cell and their spatial diffusion throughout the chamber and their effect on neighboring cells.
The team simplified the high-dimensional system into a one-dimensional orbit, noting that the system repeats periodically. This allowed them to discover that cells can make one voice when they lowered their own voice and listened to the others. “It turns out the positive feedback loop reduces the distance between moving points and finally makes them move all together. That’s why you clap louder when you hear applause from nearby neighbors and everyone eventually claps together at almost the same time,” said Professor Kim.
Professor Kim added, “Math is a powerful as it simplifies complex thing so that we can find an essential underlying property. This finding would not have been possible without the simplification of complex systems using mathematics."
The National Institutes of Health, the National Science Foundation, the Robert A. Welch Foundation, the Hamill Foundation, the National Research Foundation of Korea, and the T.J. Park Science Fellowship of POSCO supported the research.
(Figure: Complex molecular interactions among microbial consortia is simplified as interactions among points on a limit cycle (right).)
2019.10.15 View 29979 -
 Two Professors Recognized for the National R&D Excellence 100
< Professor Haeng-Ki Lee (left) and Professor Jeong-Ho Lee (right) >
Two KAIST professors were listed among the 2019 National R&D Excellence 100 announced by the Ministry of Science and ICT and the Korea Institute of S&T Evaluation and Planning.
Professor Haeng-Ki Lee from the Department of Civil and Environmental Engineering was recognized in the field of mechanics and materials for his research on developing new construction materials through the convergence of nano- and biotechnologies.
In the field of life and marine science, Professor Jeong-Ho Lee from the Graduate School of Medical Science and Engineering was lauded for his research of diagnostic tools and therapies for glioblastoma and pediatric brain tumors.
A certificate from the Minister of Ministry of Science and ICT will be conferred to these two professors, and their names will be inscribed on a special 2019 National R&D Excellence 100 plaque to celebrate their achievements. The professors will also be given privileges during the process of new R&D project selection.
(END)
2019.10.15 View 14018
Two Professors Recognized for the National R&D Excellence 100
< Professor Haeng-Ki Lee (left) and Professor Jeong-Ho Lee (right) >
Two KAIST professors were listed among the 2019 National R&D Excellence 100 announced by the Ministry of Science and ICT and the Korea Institute of S&T Evaluation and Planning.
Professor Haeng-Ki Lee from the Department of Civil and Environmental Engineering was recognized in the field of mechanics and materials for his research on developing new construction materials through the convergence of nano- and biotechnologies.
In the field of life and marine science, Professor Jeong-Ho Lee from the Graduate School of Medical Science and Engineering was lauded for his research of diagnostic tools and therapies for glioblastoma and pediatric brain tumors.
A certificate from the Minister of Ministry of Science and ICT will be conferred to these two professors, and their names will be inscribed on a special 2019 National R&D Excellence 100 plaque to celebrate their achievements. The professors will also be given privileges during the process of new R&D project selection.
(END)
2019.10.15 View 14018 -
 Professor Byong-Guk Park Named Scientist of October
< Professor Byong-Guk Park >
Professor Byong-Guk Park from the Department of Materials Science and Engineering was selected as the ‘Scientist of the Month’ for October 2019 by the Ministry of Science and ICT and the National Research Foundation of Korea. Professor Park was recognized for his contributions to the advancement of spin-orbit torque (SOT)-based magnetic random access memory (MRAM) technology. He received 10 million KRW in prize money.
A next-generation, non-volatile memory device MRAM consists of thin magnetic films. It can be applied in “logic-in-memory” devices, in which logic and memory functionalities coexist, thus drastically improving the performance of complementary metal-oxide semiconductor (CMOS) processors. Conventional MRAM technology is limited in its ability to increase the operation speed of a memory device while maintaining a high density.
Professor Park tackled this challenge by introducing a new material, antiferromagnet (IrMn), that generates a sizable amount of SOT as well as an exchange-bias field, which makes successful data writing possible without an external magnetic field. This research outcome paved the way for the development of MRAM, which has a simple device structure but features high speeds and density.
Professor Park said, “I feel rewarded to have forwarded the feasibility and applicability of MRAM. I will continue devoting myself to studying further on the development of new materials that can help enhance the performance of memory devices."
(END)
2019.10.10 View 11847
Professor Byong-Guk Park Named Scientist of October
< Professor Byong-Guk Park >
Professor Byong-Guk Park from the Department of Materials Science and Engineering was selected as the ‘Scientist of the Month’ for October 2019 by the Ministry of Science and ICT and the National Research Foundation of Korea. Professor Park was recognized for his contributions to the advancement of spin-orbit torque (SOT)-based magnetic random access memory (MRAM) technology. He received 10 million KRW in prize money.
A next-generation, non-volatile memory device MRAM consists of thin magnetic films. It can be applied in “logic-in-memory” devices, in which logic and memory functionalities coexist, thus drastically improving the performance of complementary metal-oxide semiconductor (CMOS) processors. Conventional MRAM technology is limited in its ability to increase the operation speed of a memory device while maintaining a high density.
Professor Park tackled this challenge by introducing a new material, antiferromagnet (IrMn), that generates a sizable amount of SOT as well as an exchange-bias field, which makes successful data writing possible without an external magnetic field. This research outcome paved the way for the development of MRAM, which has a simple device structure but features high speeds and density.
Professor Park said, “I feel rewarded to have forwarded the feasibility and applicability of MRAM. I will continue devoting myself to studying further on the development of new materials that can help enhance the performance of memory devices."
(END)
2019.10.10 View 11847