GE
-
 KAIST Team Develops Surface-Lighting MicroLED Patch with Significant Melanogenesis Inhibition Effect
A KAIST research team led by Ph.d candidate Jae Hee Lee and Professor Keon Jae Lee from the Department of Materials Science and Engineering has developed a surface-lighting microLED patch for UV-induced melanogenesis inhibition.
Melanin is brown or dark pigments existing in the skin, which can be abnormally synthesized by external UV or stress. Since the excessive melanin leads to skin diseases such as spots and freckles, proper treatment is required to return normal skin condition.
Recently, LED-based photo-stimulators have been released for skin care, however, their therapeutic effect is still controversial. Since conventional LED stimulators cannot conformally attach to the human skin, distance-induced side effects are caused by light loss and high heat transfer. To achieve effective phototreatment, the LED stimulator needs to be irradiated in contact with the human skin surface, enabling proper and uniform light deliver to the dermis with minimal optical loss.
In this work, the research team fabricated skin-attachable surface-lighting microLED (SµLED, 4 × 4 cm2) patch by utilizing a thousand of microLED chips and silica-embedded light diffusion layer. 100 µm-sized LED chips are vertically-interconnected for high flexibility and low heat generation, allowing its long-term operation on the human skin.
< Image 1. The overall concept of SµLED patch. a) SµLED patch operated on the human skin. b) Schematic illustration of SµLED patch structure. c) 4 × 4 cm2-sized SµLED patch. d) Schematic illustration of the advantages of SµLED patch such as efficient light delivery, low heat generation, and surface-lighting irradiation. >
The research team confirmed melanogenesis inhibition by irradiating the SµLED patch and the conventional LED (CLED) on the artificial human skin and mice dorsal skin. The SµLED-treated groups of human cells and mouse tissues showed minimal epidermal photo-toxicity and consistently effective reduction in synthesized melanin, compared to CLED-treated groups. In addition, significant suppression of proteins/catalysts expression involved in melanin synthesis such as MITF (microphthalmia-associated transcription factor), Melan-A and tyrosinase was verified.
< Image 2. The efficacy of melanogenesis inhibition on 3D human skin cells. a). Different irradiation conditions for a-MSH (major factor to stimulate melanin synthesis) treated cells. b) The ratio of pigmented area to total epidermis area. c) Relative variance of melanin level in 1 cm2-sized skin cells. A low variance means that melanin is evenly distributed, and a high variance means that the melanin is irregularly distributed. d) Optical images after in vitro experiments for 12 days. Scale bar, 1cm. e) Histological analysis of 3D skin, showing the greatest reduction in melanin after SµLED irradiation. Scale bar, 20 µm. >
< Image 3. The efficacy of melanogenesis inhibition on mouse dorsal skin. a) Optical images of mice dorsal skin after photo-treatment for 20 days. b) Histological analysis of mice dorsal skin. Less brown color means less expression of protein/catalysis involved in melanin synthesis. Scale bar, 50 µm. >
Prof. Keon Jae Lee said, “Our inorganic-based SµLED patch has outstanding characteristics in light efficiency, reliability, and durability. The SµLED patch is expected to give a great impact on the cosmetic field by reducing side effects and maximizing phototherapeutic effects.” The core technology of cosmetic SµLED has been transferred to Fronics co., Ltd, founded by Prof. Lee. Fronics is building foundry and equipment for mass production of SµLED masks for whole face cover and plans to release the products in March next year.
This paper entitled “Wearable Surface-Lighting Micro-Light-Emitting Diode Patch for Melanogenesis Inhibition” was published in the November 2022 issue of Advanced Healthcare Materials.
2022.11.22 View 13049
KAIST Team Develops Surface-Lighting MicroLED Patch with Significant Melanogenesis Inhibition Effect
A KAIST research team led by Ph.d candidate Jae Hee Lee and Professor Keon Jae Lee from the Department of Materials Science and Engineering has developed a surface-lighting microLED patch for UV-induced melanogenesis inhibition.
Melanin is brown or dark pigments existing in the skin, which can be abnormally synthesized by external UV or stress. Since the excessive melanin leads to skin diseases such as spots and freckles, proper treatment is required to return normal skin condition.
Recently, LED-based photo-stimulators have been released for skin care, however, their therapeutic effect is still controversial. Since conventional LED stimulators cannot conformally attach to the human skin, distance-induced side effects are caused by light loss and high heat transfer. To achieve effective phototreatment, the LED stimulator needs to be irradiated in contact with the human skin surface, enabling proper and uniform light deliver to the dermis with minimal optical loss.
In this work, the research team fabricated skin-attachable surface-lighting microLED (SµLED, 4 × 4 cm2) patch by utilizing a thousand of microLED chips and silica-embedded light diffusion layer. 100 µm-sized LED chips are vertically-interconnected for high flexibility and low heat generation, allowing its long-term operation on the human skin.
< Image 1. The overall concept of SµLED patch. a) SµLED patch operated on the human skin. b) Schematic illustration of SµLED patch structure. c) 4 × 4 cm2-sized SµLED patch. d) Schematic illustration of the advantages of SµLED patch such as efficient light delivery, low heat generation, and surface-lighting irradiation. >
The research team confirmed melanogenesis inhibition by irradiating the SµLED patch and the conventional LED (CLED) on the artificial human skin and mice dorsal skin. The SµLED-treated groups of human cells and mouse tissues showed minimal epidermal photo-toxicity and consistently effective reduction in synthesized melanin, compared to CLED-treated groups. In addition, significant suppression of proteins/catalysts expression involved in melanin synthesis such as MITF (microphthalmia-associated transcription factor), Melan-A and tyrosinase was verified.
< Image 2. The efficacy of melanogenesis inhibition on 3D human skin cells. a). Different irradiation conditions for a-MSH (major factor to stimulate melanin synthesis) treated cells. b) The ratio of pigmented area to total epidermis area. c) Relative variance of melanin level in 1 cm2-sized skin cells. A low variance means that melanin is evenly distributed, and a high variance means that the melanin is irregularly distributed. d) Optical images after in vitro experiments for 12 days. Scale bar, 1cm. e) Histological analysis of 3D skin, showing the greatest reduction in melanin after SµLED irradiation. Scale bar, 20 µm. >
< Image 3. The efficacy of melanogenesis inhibition on mouse dorsal skin. a) Optical images of mice dorsal skin after photo-treatment for 20 days. b) Histological analysis of mice dorsal skin. Less brown color means less expression of protein/catalysis involved in melanin synthesis. Scale bar, 50 µm. >
Prof. Keon Jae Lee said, “Our inorganic-based SµLED patch has outstanding characteristics in light efficiency, reliability, and durability. The SµLED patch is expected to give a great impact on the cosmetic field by reducing side effects and maximizing phototherapeutic effects.” The core technology of cosmetic SµLED has been transferred to Fronics co., Ltd, founded by Prof. Lee. Fronics is building foundry and equipment for mass production of SµLED masks for whole face cover and plans to release the products in March next year.
This paper entitled “Wearable Surface-Lighting Micro-Light-Emitting Diode Patch for Melanogenesis Inhibition” was published in the November 2022 issue of Advanced Healthcare Materials.
2022.11.22 View 13049 -
 See-through exhibitions using smartphones: KAIST develops the AR magic lens, WonderScope
WonderScope shows what’s underneath the surface of an object through an augmented reality technology.
< Photo 1. Demonstration at ACM SIGGRAPH >
- A KAIST research team led by Professor Woohun Lee from the Department of Industrial Design and Professor Geehyuk Lee from the School of Computing have developed a smartphone “appcessory” called WonderScope that can easily add an augmented reality (AR) perspective to the surface of exhibits
- The research won an Honorable Mention for Emerging Technologies Best in Show at ACM SIGGRAPH, one of the largest international conferences on computer graphics and interactions
- The technology was improved and validated through real-life applications in three special exhibitions including one at the Geological Museum at the Korea Institute of Geoscience and Mineral Resources (KIGAM) held in 2020, and two at the National Science Museum each in 2021 and 2022
- The technology is expected to be used for public science exhibitions and museums as well as for interactive teaching materials to stimulate children’s curiosity
A KAIST research team led by Professor Woohun Lee from the Department of Industrial Design and Professor Geehyuk Lee from the School of Computing developed a novel augmented reality (AR) device, WonderScope, which displays the insides of an object directly from its surface. By installing and connecting WonderScope to a mobile device through Bluetooth, users can see through exhibits as if looking through a magic lens.
Many science museums nowadays have incorporated the use of AR apps for mobile devices. Such apps add digital information to the exhibition, providing a unique experience. However, visitors must watch the screen from a certain distance away from the exhibited items, often causing them to focus more on the digital contents rather than the exhibits themselves. In other words, the distance and distractions that exist between the exhibit and the mobile device may actually cause the visitors to feel detached from the exhibition. To solve this problem, museums needed a magic AR lens that could be used directly from the surface of the item.
To accomplish this, smartphones must know exactly where on the surface of an object it is placed. Generally, this would require an additional recognition device either on the inside or on the surface of the item, or a special pattern printed on its surface. Realistically speaking, these are impractical solutions, as exhibits would either appear overly complex or face spatial restrictions.
WonderScope, on the other hand, uses a much more practical method to identify the location of a smartphone on the surface of an exhibit. First, it reads a small RFID tag attached to the surface of an object, and calculates the location of the moving smartphone by adding its relative movements based on the readings from an optical displacement sensor and an acceleration sensor. The research team also took into consideration the height of the smartphone, and the characteristics of the surface profile in order to calculate the device’s position more accurately. By attaching or embedding RFID tags on exhibits, visitors can easily experience the effects of a magic AR lens through their smartphones.
For its wider use, WonderScope must be able to locate itself from various types of exhibit surfaces. To this end, WoderScope uses readings from an optical displacement sensor and an acceleration sensor with complementary characteristics, allowing stable locating capacities on various textures including paper, stone, wood, plastic, acrylic, and glass, as well as surfaces with physical patterns or irregularities. As a result, WonderScope can identify its location from a distance as close as 4 centimeters from an object, also enabling simple three-dimensional interactions near the surface of the exhibits.
The research team developed various case project templates and WonderScope support tools to allow the facile production of smartphone apps that use general-purpose virtual reality (VR) and the game engine Unity. WonderScope is also compatible with various types of devices that run on the Android operating system, including smartwatches, smartphones, and tablets, allowing it to be applied to exhibitions in many forms.
< Photo 2. Human body model showing demonstration >
< Photo 3. Demonstration of the underground mineral exploration game >
< Photo 4. Demonstration of Apollo 11 moon exploration experience >
The research team developed WonderScope with funding from the science and culture exhibition enhancement support project by the Ministry of Science and ICT. Between October 27, 2020 and February 28, 2021, WonderScope was used to observe underground volcanic activity and the insides of volcanic rocks at “There Once was a Volcano”, a special exhibition held at the Geological Museum in the Korea institute of Geoscience and Mineral Resources (KIGAM). From September 28 to October 3, 2021, it was used to observe the surface of Jung-moon-kyung (a bronze mirror with fine linear design) at the special exhibition “A Bronze Mirror Shines on Science” at the National Science Museum. And from August 2 to October 3, 2022 it was applied to a moon landing simulation at “The Special Exhibition on Moon Exploration”, also at the National Science Museum. Through various field demonstrations over the years, the research team has improved the performance and usability of WonderScope.
< Photo 5. Observation of surface corrosion of the main gate >
The research team demonstrated WonderScope at the Emerging Technologies forum during ACM SIGGRAPH 2022, a computer graphics and interaction technology conference that was held in Vancouver, Canada between August 8 and 11 this year. At this conference, where the latest interactive technologies are introduced, the team won an Honorable Mention for Best in Show. The judges commented that “WonderScope will be a new technology that provides the audience with a unique joy of participation during their visits to exhibitions and museums.”
< Photo 6. Cover of Digital Creativity >
WonderScope is a cylindrical “appcessory” module, 5cm in diameter and 4.5cm in height. It is small enough to be easily attached to a smartphone and embedded on most exhibits. Professor Woohun Lee from the KAIST Department of Industrial Design, who supervised the research, said, “WonderScope can be applied to various applications including not only educational, but also industrial exhibitions, in many ways.” He added, “We also expect for it to be used as an interactive teaching tool that stimulates children’s curiosity.”
Introductory video of WonderScope: https://www.youtube.com/watch?v=X2MyAXRt7h4&t=7s
2022.10.24 View 11189
See-through exhibitions using smartphones: KAIST develops the AR magic lens, WonderScope
WonderScope shows what’s underneath the surface of an object through an augmented reality technology.
< Photo 1. Demonstration at ACM SIGGRAPH >
- A KAIST research team led by Professor Woohun Lee from the Department of Industrial Design and Professor Geehyuk Lee from the School of Computing have developed a smartphone “appcessory” called WonderScope that can easily add an augmented reality (AR) perspective to the surface of exhibits
- The research won an Honorable Mention for Emerging Technologies Best in Show at ACM SIGGRAPH, one of the largest international conferences on computer graphics and interactions
- The technology was improved and validated through real-life applications in three special exhibitions including one at the Geological Museum at the Korea Institute of Geoscience and Mineral Resources (KIGAM) held in 2020, and two at the National Science Museum each in 2021 and 2022
- The technology is expected to be used for public science exhibitions and museums as well as for interactive teaching materials to stimulate children’s curiosity
A KAIST research team led by Professor Woohun Lee from the Department of Industrial Design and Professor Geehyuk Lee from the School of Computing developed a novel augmented reality (AR) device, WonderScope, which displays the insides of an object directly from its surface. By installing and connecting WonderScope to a mobile device through Bluetooth, users can see through exhibits as if looking through a magic lens.
Many science museums nowadays have incorporated the use of AR apps for mobile devices. Such apps add digital information to the exhibition, providing a unique experience. However, visitors must watch the screen from a certain distance away from the exhibited items, often causing them to focus more on the digital contents rather than the exhibits themselves. In other words, the distance and distractions that exist between the exhibit and the mobile device may actually cause the visitors to feel detached from the exhibition. To solve this problem, museums needed a magic AR lens that could be used directly from the surface of the item.
To accomplish this, smartphones must know exactly where on the surface of an object it is placed. Generally, this would require an additional recognition device either on the inside or on the surface of the item, or a special pattern printed on its surface. Realistically speaking, these are impractical solutions, as exhibits would either appear overly complex or face spatial restrictions.
WonderScope, on the other hand, uses a much more practical method to identify the location of a smartphone on the surface of an exhibit. First, it reads a small RFID tag attached to the surface of an object, and calculates the location of the moving smartphone by adding its relative movements based on the readings from an optical displacement sensor and an acceleration sensor. The research team also took into consideration the height of the smartphone, and the characteristics of the surface profile in order to calculate the device’s position more accurately. By attaching or embedding RFID tags on exhibits, visitors can easily experience the effects of a magic AR lens through their smartphones.
For its wider use, WonderScope must be able to locate itself from various types of exhibit surfaces. To this end, WoderScope uses readings from an optical displacement sensor and an acceleration sensor with complementary characteristics, allowing stable locating capacities on various textures including paper, stone, wood, plastic, acrylic, and glass, as well as surfaces with physical patterns or irregularities. As a result, WonderScope can identify its location from a distance as close as 4 centimeters from an object, also enabling simple three-dimensional interactions near the surface of the exhibits.
The research team developed various case project templates and WonderScope support tools to allow the facile production of smartphone apps that use general-purpose virtual reality (VR) and the game engine Unity. WonderScope is also compatible with various types of devices that run on the Android operating system, including smartwatches, smartphones, and tablets, allowing it to be applied to exhibitions in many forms.
< Photo 2. Human body model showing demonstration >
< Photo 3. Demonstration of the underground mineral exploration game >
< Photo 4. Demonstration of Apollo 11 moon exploration experience >
The research team developed WonderScope with funding from the science and culture exhibition enhancement support project by the Ministry of Science and ICT. Between October 27, 2020 and February 28, 2021, WonderScope was used to observe underground volcanic activity and the insides of volcanic rocks at “There Once was a Volcano”, a special exhibition held at the Geological Museum in the Korea institute of Geoscience and Mineral Resources (KIGAM). From September 28 to October 3, 2021, it was used to observe the surface of Jung-moon-kyung (a bronze mirror with fine linear design) at the special exhibition “A Bronze Mirror Shines on Science” at the National Science Museum. And from August 2 to October 3, 2022 it was applied to a moon landing simulation at “The Special Exhibition on Moon Exploration”, also at the National Science Museum. Through various field demonstrations over the years, the research team has improved the performance and usability of WonderScope.
< Photo 5. Observation of surface corrosion of the main gate >
The research team demonstrated WonderScope at the Emerging Technologies forum during ACM SIGGRAPH 2022, a computer graphics and interaction technology conference that was held in Vancouver, Canada between August 8 and 11 this year. At this conference, where the latest interactive technologies are introduced, the team won an Honorable Mention for Best in Show. The judges commented that “WonderScope will be a new technology that provides the audience with a unique joy of participation during their visits to exhibitions and museums.”
< Photo 6. Cover of Digital Creativity >
WonderScope is a cylindrical “appcessory” module, 5cm in diameter and 4.5cm in height. It is small enough to be easily attached to a smartphone and embedded on most exhibits. Professor Woohun Lee from the KAIST Department of Industrial Design, who supervised the research, said, “WonderScope can be applied to various applications including not only educational, but also industrial exhibitions, in many ways.” He added, “We also expect for it to be used as an interactive teaching tool that stimulates children’s curiosity.”
Introductory video of WonderScope: https://www.youtube.com/watch?v=X2MyAXRt7h4&t=7s
2022.10.24 View 11189 -
 Globally renowned stained-glass artist Fr. En Joon Kim appointed as a distinguished invited professor in the KAIST Department of Industrial Design
World-renowned master of stained-glass Father En Joong Kim was appointed to a two-year distinguished invited professorship in the KAIST Department of Industrial Design starting August 1, 2022
- Fr. Kim will share his life, spirit, and artistic capabilities with the members of KAIST through special lectures for undergraduate and graduate students, and through a stained-glass piece he will work on and donate to the KAIST Academic and Cultural Complex
- The 53-piece work of art will provide KAIST with fresh inspiration and add to its dynamic atmosphere
KAIST appointed the world-renowned stained-glass artist and priest Fr. En Joong Kim of the Dominican Order as a distinguished invited professor in the KAIST Department of Industrial Design. His term starts from August 1 of this year and ends on July 31, 2024.
The appointment aims to share the life, spirit, and artistic capabilities of Fr. Kim, who is internationally recognized for his creative work. The purpose of the appointment is not only to provide professional advice on lighting color and space, which are core contents of industrial design courses, but also to bring new inspiration to KAIST community.
Fr. Kim, who studied in the College of Fine Arts at Seoul National University, won the Korean Art Award in 1965, and later studied at the University of Fribourg in Switzerland and the Catholic University of Paris. Joining the Dominican Order in France in 1974, he started his career as both a priest and an artist, and continued his artistic activities via 200 exhibitions around the world and by working on the stained-glass windows of 50 European churches.
In recognition of the artistic merit of combining colorful tones with the beauty of blank spaces, a distinctive characteristic of Asian art, and Fr. Kim’s contributions to establishing such combinations, Passage Kim En Joong, an art gallery, was founded in Ambert, France in 2019, and for his artwork installed all over France, he was presented with the insignia of Officer in the Order of Arts and Letters by the French government in 2010.
Following the appointment, the KAIST Department of Industrial Design is preparing a special seminar lecture by Fr. Kim under the title “Search the Future”. Fr. Kim will share his experience and philosophy for pursuing aesthetic values and efforts. In addition, the department plans to set up a special studio for Fr. Kim to both work and interact with students, encouraging them to naturally communicate and share ideas together.
One of Fr. Kim's art piece being installed at the main administration building at KAIST.
In a studio at the KAIST Academic Cultural Complex (ACC), Fr. Kim is currently working on his 53-piece stained-glass project that, when finished, will be added to the ACC. KAISTians will be able to enjoy a master’s art on a daily basis as the 53 sheets of glasses combine to form one magnificent piece.
Fr. Kim said, “I am very happy to be a distinguished invited professor at KAIST, where excellent scientists are at work. It is my wish and prayer that my presence here may comfort the students’ hearts with artwork and art philosophy that carries sensitivity and sincerity, and that they may garner richer experiences.”
KAIST President Kwang Hyung Lee said, “The purpose of research and art are similar in that they pioneer through endless contemplations and attempts. The art piece to be installed at ACC, which will combine 53 pieces of stained glass, resembles our school, where our members each with their own distinctive colors and textures come together create a harmonious new form known as KAIST.”
He added, “I hope that the artistic spirit of Fr. Kim, a world-class master, will be a beacon that would bring a new type of stimulation and ease here at KAIST”
KAIST also appointed world-renowned soprano Sumi Jo as a distinguished invited professor in the Graduate School of Culture Technology in October 2021, and SM Entertainment’s executive producer Soo-man Lee as a distinguished invited professor in the School of Computing in March 2022. KAIST continues to expand and incorporate science and technology into the fields of art and culture, and to establish itself as a place for joint research and creative endeavors.
2022.09.08 View 10623
Globally renowned stained-glass artist Fr. En Joon Kim appointed as a distinguished invited professor in the KAIST Department of Industrial Design
World-renowned master of stained-glass Father En Joong Kim was appointed to a two-year distinguished invited professorship in the KAIST Department of Industrial Design starting August 1, 2022
- Fr. Kim will share his life, spirit, and artistic capabilities with the members of KAIST through special lectures for undergraduate and graduate students, and through a stained-glass piece he will work on and donate to the KAIST Academic and Cultural Complex
- The 53-piece work of art will provide KAIST with fresh inspiration and add to its dynamic atmosphere
KAIST appointed the world-renowned stained-glass artist and priest Fr. En Joong Kim of the Dominican Order as a distinguished invited professor in the KAIST Department of Industrial Design. His term starts from August 1 of this year and ends on July 31, 2024.
The appointment aims to share the life, spirit, and artistic capabilities of Fr. Kim, who is internationally recognized for his creative work. The purpose of the appointment is not only to provide professional advice on lighting color and space, which are core contents of industrial design courses, but also to bring new inspiration to KAIST community.
Fr. Kim, who studied in the College of Fine Arts at Seoul National University, won the Korean Art Award in 1965, and later studied at the University of Fribourg in Switzerland and the Catholic University of Paris. Joining the Dominican Order in France in 1974, he started his career as both a priest and an artist, and continued his artistic activities via 200 exhibitions around the world and by working on the stained-glass windows of 50 European churches.
In recognition of the artistic merit of combining colorful tones with the beauty of blank spaces, a distinctive characteristic of Asian art, and Fr. Kim’s contributions to establishing such combinations, Passage Kim En Joong, an art gallery, was founded in Ambert, France in 2019, and for his artwork installed all over France, he was presented with the insignia of Officer in the Order of Arts and Letters by the French government in 2010.
Following the appointment, the KAIST Department of Industrial Design is preparing a special seminar lecture by Fr. Kim under the title “Search the Future”. Fr. Kim will share his experience and philosophy for pursuing aesthetic values and efforts. In addition, the department plans to set up a special studio for Fr. Kim to both work and interact with students, encouraging them to naturally communicate and share ideas together.
One of Fr. Kim's art piece being installed at the main administration building at KAIST.
In a studio at the KAIST Academic Cultural Complex (ACC), Fr. Kim is currently working on his 53-piece stained-glass project that, when finished, will be added to the ACC. KAISTians will be able to enjoy a master’s art on a daily basis as the 53 sheets of glasses combine to form one magnificent piece.
Fr. Kim said, “I am very happy to be a distinguished invited professor at KAIST, where excellent scientists are at work. It is my wish and prayer that my presence here may comfort the students’ hearts with artwork and art philosophy that carries sensitivity and sincerity, and that they may garner richer experiences.”
KAIST President Kwang Hyung Lee said, “The purpose of research and art are similar in that they pioneer through endless contemplations and attempts. The art piece to be installed at ACC, which will combine 53 pieces of stained glass, resembles our school, where our members each with their own distinctive colors and textures come together create a harmonious new form known as KAIST.”
He added, “I hope that the artistic spirit of Fr. Kim, a world-class master, will be a beacon that would bring a new type of stimulation and ease here at KAIST”
KAIST also appointed world-renowned soprano Sumi Jo as a distinguished invited professor in the Graduate School of Culture Technology in October 2021, and SM Entertainment’s executive producer Soo-man Lee as a distinguished invited professor in the School of Computing in March 2022. KAIST continues to expand and incorporate science and technology into the fields of art and culture, and to establish itself as a place for joint research and creative endeavors.
2022.09.08 View 10623 -
 A KAIST Research Team Develops Diesel Reforming Catalyst Enabling Hydrogen Production for Future Mobile Fuel Cells
This catalyst capability allowing stable hydrogen production from commercial diesel is expected to be applied in mobile fuel cell systems in the future hydrogen economy
On August 16, a joint research team led by Professors Joongmyeon Bae and Kang Taek Lee of KAIST’s Department of Mechanical Engineering and Dr. Chan-Woo Lee of Korea Institute of Energy Research (KIER) announced the successful development of a highly active and durable reforming catalyst allowing hydrogen production from commercial diesel.
Fuel reforming is a hydrogen production technique that extracts hydrogen from hydrocarbons through catalytic reactions. Diesel, being a liquid fuel, has a high storage density for hydrogen and is easy to transport and store. There have therefore been continuous research efforts to apply hydrogel supply systems using diesel reformation in mobile fuel cells, such as for auxiliary power in heavy trucks or air-independent propulsion (AIP) systems in submarines.
However, diesel is a mixture of high hydrocarbons including long-chained paraffin, double-bonded olefin, and aromatic hydrocarbons with benzene groups, and it requires a highly active catalyst to effectively break them down. In addition, the catalyst must be extremely durable against caulking and sintering, as they are often the main causes of catalyst degradation. Such challenges have limited the use of diesel reformation technologies to date.
The joint research team successfully developed a highly active and durable diesel reforming catalyst through elution (a heat treatment method used to uniformly grow active metals retained in an oxide support as ions in the form of metal nanoparticles), forming alloy nanoparticles. The design was based on the fact that eluted nanoparticles strongly interact with the support, allowing a high degree of dispersion at high temperatures, and that producing an alloy from dissimilar metals can increase the performance of catalysts through a synergistic effect.
The research team introduced a solution combustion synthesis method to produce a multi-component catalyst with a trace amount of platinum (Pt) and ruthenium (Ru) penetrated into a ceria (CeO2) lattice, which is a structure commonly used as a support for catalysts in redox reactions. When exposed to a diesel reforming reaction environment, the catalyst induces Pt-Ru alloy nanoparticle formation upon Pt and Ru elution onto the support surface.
In addition to the catalyst analysis, the research team also succeeded in characterizing the behaviour of active metal elution and alloy formation from an energetic perspective using a density functional theory-based calculation. In a performance comparison test between the Pt-Ru alloy catalyst against existing single-metal catalysts, the reforming activity was shown to have improved, as it showed a 100% fuel conversion rate even at a low temperature (600oC, compared to the original 800oC). In a long-term durability test (800oC, 200 hours), the catalyst showed commercial stability by successfully producing hydrogen from commercial diesel without performance degradation.
The study was conducted by Ph.D. candidate Jaemyung Lee of KAIST’s Department of Mechanical Engineering as the first author. Ph.D. candidate Changho Yeon of KIER, Dr. Jiwoo Oh of KAIST’s Department of Mechanical Engineering, Dr. Gwangwoo Han of KIER, Ph.D. candidate Jeong Do Yoo of KAIST’s Department of Mechanical Engineering, and Dr. Hyung Joong Yun of the Korea Basic Science Institute contributed as co-authors. Dr. Chan-Woo Lee of KIER and Professors Kang Taek Lee and Joongmyeon Bae of KAIST’s Department of Mechanical Engineering contributed as corresponding authors. The research was published in the online version of Applied Catalysis B: Environmental (IF 24.319, JCR 0.93%) on June 17, under the title “Highly Active and Stable Catalyst with Exsolved PtRu Alloy Nanoparticles for Hydrogen Production via Commercial Diesel Reforming”.
Professor Joongmyeon Bae said, “The fact that hydrogen can be stably produced from commercial diesel makes this a very meaningful achievement, and we look forward to this technology contributing to the active introduction of mobile fuel cell systems in the early hydrogen economy.” He added, “Our approach to catalyst design may be applied not only to reforming reactions, but also in various other fields.”
This research was supported by the National Research Foundation of Korea through funding from the Ministry of Science, ICT and Future Planning.
Figure. Schematic diagram of high-performance diesel reforming catalyst with eluted platinum-ruthenium alloy nanoparticles and long-term durability verification experiment results for commercial diesel reforming reaction
2022.09.07 View 15239
A KAIST Research Team Develops Diesel Reforming Catalyst Enabling Hydrogen Production for Future Mobile Fuel Cells
This catalyst capability allowing stable hydrogen production from commercial diesel is expected to be applied in mobile fuel cell systems in the future hydrogen economy
On August 16, a joint research team led by Professors Joongmyeon Bae and Kang Taek Lee of KAIST’s Department of Mechanical Engineering and Dr. Chan-Woo Lee of Korea Institute of Energy Research (KIER) announced the successful development of a highly active and durable reforming catalyst allowing hydrogen production from commercial diesel.
Fuel reforming is a hydrogen production technique that extracts hydrogen from hydrocarbons through catalytic reactions. Diesel, being a liquid fuel, has a high storage density for hydrogen and is easy to transport and store. There have therefore been continuous research efforts to apply hydrogel supply systems using diesel reformation in mobile fuel cells, such as for auxiliary power in heavy trucks or air-independent propulsion (AIP) systems in submarines.
However, diesel is a mixture of high hydrocarbons including long-chained paraffin, double-bonded olefin, and aromatic hydrocarbons with benzene groups, and it requires a highly active catalyst to effectively break them down. In addition, the catalyst must be extremely durable against caulking and sintering, as they are often the main causes of catalyst degradation. Such challenges have limited the use of diesel reformation technologies to date.
The joint research team successfully developed a highly active and durable diesel reforming catalyst through elution (a heat treatment method used to uniformly grow active metals retained in an oxide support as ions in the form of metal nanoparticles), forming alloy nanoparticles. The design was based on the fact that eluted nanoparticles strongly interact with the support, allowing a high degree of dispersion at high temperatures, and that producing an alloy from dissimilar metals can increase the performance of catalysts through a synergistic effect.
The research team introduced a solution combustion synthesis method to produce a multi-component catalyst with a trace amount of platinum (Pt) and ruthenium (Ru) penetrated into a ceria (CeO2) lattice, which is a structure commonly used as a support for catalysts in redox reactions. When exposed to a diesel reforming reaction environment, the catalyst induces Pt-Ru alloy nanoparticle formation upon Pt and Ru elution onto the support surface.
In addition to the catalyst analysis, the research team also succeeded in characterizing the behaviour of active metal elution and alloy formation from an energetic perspective using a density functional theory-based calculation. In a performance comparison test between the Pt-Ru alloy catalyst against existing single-metal catalysts, the reforming activity was shown to have improved, as it showed a 100% fuel conversion rate even at a low temperature (600oC, compared to the original 800oC). In a long-term durability test (800oC, 200 hours), the catalyst showed commercial stability by successfully producing hydrogen from commercial diesel without performance degradation.
The study was conducted by Ph.D. candidate Jaemyung Lee of KAIST’s Department of Mechanical Engineering as the first author. Ph.D. candidate Changho Yeon of KIER, Dr. Jiwoo Oh of KAIST’s Department of Mechanical Engineering, Dr. Gwangwoo Han of KIER, Ph.D. candidate Jeong Do Yoo of KAIST’s Department of Mechanical Engineering, and Dr. Hyung Joong Yun of the Korea Basic Science Institute contributed as co-authors. Dr. Chan-Woo Lee of KIER and Professors Kang Taek Lee and Joongmyeon Bae of KAIST’s Department of Mechanical Engineering contributed as corresponding authors. The research was published in the online version of Applied Catalysis B: Environmental (IF 24.319, JCR 0.93%) on June 17, under the title “Highly Active and Stable Catalyst with Exsolved PtRu Alloy Nanoparticles for Hydrogen Production via Commercial Diesel Reforming”.
Professor Joongmyeon Bae said, “The fact that hydrogen can be stably produced from commercial diesel makes this a very meaningful achievement, and we look forward to this technology contributing to the active introduction of mobile fuel cell systems in the early hydrogen economy.” He added, “Our approach to catalyst design may be applied not only to reforming reactions, but also in various other fields.”
This research was supported by the National Research Foundation of Korea through funding from the Ministry of Science, ICT and Future Planning.
Figure. Schematic diagram of high-performance diesel reforming catalyst with eluted platinum-ruthenium alloy nanoparticles and long-term durability verification experiment results for commercial diesel reforming reaction
2022.09.07 View 15239 -
 Establishing a novel strategy to tackle Huntington’s disease
A platform to take on the Huntington’s disease via an innovative approach established by KAIST’s researchers through international collaboration with scientists in the Netherlands, France, and Sweden.
Through an international joint research effort involving ProQR Therapeutics of the Netherlands, Université Grenoble Alpes of France, and KTH Royal Institute of Technology of Sweden, Professor Ji-Soon Song's research team in the Department of Biological Sciences and KAIST Institute for BioCentury of KAIST, established a noble strategy to treat Huntington's disease. The new works showed that the protein converted from disease form to its disease-free form maintains its original function, providing new roadblocks to approach Huntington’s disease.
This research, titled, “A pathogenic-proteolysis resistant huntingtin isoform induced by an antisense oligonucleotide maintains huntingtin function”, co-authored by Hyeongju Kim, was published in the online edition of 'Journal of Clinical Investigation Insight' on August 9, 2022.
Huntington's disease is a dominantly inherited neurodegenerative disease and is caused by a mutation in a protein called ‘huntingtin’, which adds a distinctive feature of an expanded stretch of glutamine amino acids called polyglutamine to the protein. It is estimated that one in every 10,000 have Huntington's disease in United States. The patients would suffer a decade of regression before death, and, thus far, there is no known cure for the disease.
The cleavage near the stretched polyglutamine in mutated huntingtin is known to be the cause of the Huntington’s disease. However, as huntingtin protein is required for the development and normal function of the brain, it is critical to specifically eliminate the disease-causing protein while maintaining the ones that are still normally functioning.
The research team showed that huntingtin delta 12, the converted form of huntingtin that is resistant to developing cleavages at the ends of the protein, the known cause of the Huntington’s disease (HD), alleviated the disease’s symptoms while maintaining the functions of normal huntingtin.
Figure. Huntington's disease resistance huntingtin protein induced by antisense oligonucleotide (AON) is resistant to Caspase-6 cleavage, therefore, does not cause Huntington’s disease while maintaining normal functions of huntingtin.
The research was welcomed as it is sure to fuel innovate strategies to tackle Huntington’s disease without altering the essential function of huntingtin.
This work was supported by a Global Research Lab grant from the National Research Foundation of Korea (NRF) and by a EUREKA Eurostars 2 grant from European Union Horizon 2020.
2022.09.02 View 8043
Establishing a novel strategy to tackle Huntington’s disease
A platform to take on the Huntington’s disease via an innovative approach established by KAIST’s researchers through international collaboration with scientists in the Netherlands, France, and Sweden.
Through an international joint research effort involving ProQR Therapeutics of the Netherlands, Université Grenoble Alpes of France, and KTH Royal Institute of Technology of Sweden, Professor Ji-Soon Song's research team in the Department of Biological Sciences and KAIST Institute for BioCentury of KAIST, established a noble strategy to treat Huntington's disease. The new works showed that the protein converted from disease form to its disease-free form maintains its original function, providing new roadblocks to approach Huntington’s disease.
This research, titled, “A pathogenic-proteolysis resistant huntingtin isoform induced by an antisense oligonucleotide maintains huntingtin function”, co-authored by Hyeongju Kim, was published in the online edition of 'Journal of Clinical Investigation Insight' on August 9, 2022.
Huntington's disease is a dominantly inherited neurodegenerative disease and is caused by a mutation in a protein called ‘huntingtin’, which adds a distinctive feature of an expanded stretch of glutamine amino acids called polyglutamine to the protein. It is estimated that one in every 10,000 have Huntington's disease in United States. The patients would suffer a decade of regression before death, and, thus far, there is no known cure for the disease.
The cleavage near the stretched polyglutamine in mutated huntingtin is known to be the cause of the Huntington’s disease. However, as huntingtin protein is required for the development and normal function of the brain, it is critical to specifically eliminate the disease-causing protein while maintaining the ones that are still normally functioning.
The research team showed that huntingtin delta 12, the converted form of huntingtin that is resistant to developing cleavages at the ends of the protein, the known cause of the Huntington’s disease (HD), alleviated the disease’s symptoms while maintaining the functions of normal huntingtin.
Figure. Huntington's disease resistance huntingtin protein induced by antisense oligonucleotide (AON) is resistant to Caspase-6 cleavage, therefore, does not cause Huntington’s disease while maintaining normal functions of huntingtin.
The research was welcomed as it is sure to fuel innovate strategies to tackle Huntington’s disease without altering the essential function of huntingtin.
This work was supported by a Global Research Lab grant from the National Research Foundation of Korea (NRF) and by a EUREKA Eurostars 2 grant from European Union Horizon 2020.
2022.09.02 View 8043 -
 Phage resistant Escherichia coli strains developed to reduce fermentation failure
A genome engineering-based systematic strategy for developing phage resistant Escherichia coli strains has been successfully developed through the collaborative efforts of a team led by Professor Sang Yup Lee, Professor Shi Chen, and Professor Lianrong Wang. This study by Xuan Zou et al. was published in Nature Communications in August 2022 and featured in Nature Communications Editors’ Highlights. The collaboration by the School of Pharmaceutical Sciences at Wuhan University, the First Affiliated Hospital of Shenzhen University, and the KAIST Department of Chemical and Biomolecular Engineering has made an important advance in the metabolic engineering and fermentation industry as it solves a big problem of phage infection causing fermentation failure.
Systems metabolic engineering is a highly interdisciplinary field that has made the development of microbial cell factories to produce various bioproducts including chemicals, fuels, and materials possible in a sustainable and environmentally friendly way, mitigating the impact of worldwide resource depletion and climate change. Escherichia coli is one of the most important chassis microbial strains, given its wide applications in the bio-based production of a diverse range of chemicals and materials. With the development of tools and strategies for systems metabolic engineering using E. coli, a highly optimized and well-characterized cell factory will play a crucial role in converting cheap and readily available raw materials into products of great economic and industrial value.
However, the consistent problem of phage contamination in fermentation imposes a devastating impact on host cells and threatens the productivity of bacterial bioprocesses in biotechnology facilities, which can lead to widespread fermentation failure and immeasurable economic loss. Host-controlled defense systems can be developed into effective genetic engineering solutions to address bacteriophage contamination in industrial-scale fermentation; however, most of the resistance mechanisms only narrowly restrict phages and their effect on phage contamination will be limited.
There have been attempts to develop diverse abilities/systems for environmental adaptation or antiviral defense. The team’s collaborative efforts developed a new type II single-stranded DNA phosphorothioation (Ssp) defense system derived from E. coli 3234/A, which can be used in multiple industrial E. coli strains (e.g., E. coli K-12, B and W) to provide broad protection against various types of dsDNA coliphages. Furthermore, they developed a systematic genome engineering strategy involving the simultaneous genomic integration of the Ssp defense module and mutations in components that are essential to the phage life cycle. This strategy can be used to transform E. coli hosts that are highly susceptible to phage attack into strains with powerful restriction effects on the tested bacteriophages. This endows hosts with strong resistance against a wide spectrum of phage infections without affecting bacterial growth and normal physiological function. More importantly, the resulting engineered phage-resistant strains maintained the capabilities of producing the desired chemicals and recombinant proteins even under high levels of phage cocktail challenge, which provides crucial protection against phage attacks.
This is a major step forward, as it provides a systematic solution for engineering phage-resistant bacterial strains, especially industrial bioproduction strains, to protect cells from a wide range of bacteriophages. Considering the functionality of this engineering strategy with diverse E. coli strains, the strategy reported in this study can be widely extended to other bacterial species and industrial applications, which will be of great interest to researchers in academia and industry alike.
Fig. A schematic model of the systematic strategy for engineering phage-sensitive industrial E. coli strains into strains with broad antiphage activities. Through the simultaneous genomic integration of a DNA phosphorothioation-based Ssp defense module and mutations of components essential for the phage life cycle, the engineered E. coli strains show strong resistance against diverse phages tested and maintain the capabilities of producing example recombinant proteins, even under high levels of phage cocktail challenge.
2022.08.23 View 15172
Phage resistant Escherichia coli strains developed to reduce fermentation failure
A genome engineering-based systematic strategy for developing phage resistant Escherichia coli strains has been successfully developed through the collaborative efforts of a team led by Professor Sang Yup Lee, Professor Shi Chen, and Professor Lianrong Wang. This study by Xuan Zou et al. was published in Nature Communications in August 2022 and featured in Nature Communications Editors’ Highlights. The collaboration by the School of Pharmaceutical Sciences at Wuhan University, the First Affiliated Hospital of Shenzhen University, and the KAIST Department of Chemical and Biomolecular Engineering has made an important advance in the metabolic engineering and fermentation industry as it solves a big problem of phage infection causing fermentation failure.
Systems metabolic engineering is a highly interdisciplinary field that has made the development of microbial cell factories to produce various bioproducts including chemicals, fuels, and materials possible in a sustainable and environmentally friendly way, mitigating the impact of worldwide resource depletion and climate change. Escherichia coli is one of the most important chassis microbial strains, given its wide applications in the bio-based production of a diverse range of chemicals and materials. With the development of tools and strategies for systems metabolic engineering using E. coli, a highly optimized and well-characterized cell factory will play a crucial role in converting cheap and readily available raw materials into products of great economic and industrial value.
However, the consistent problem of phage contamination in fermentation imposes a devastating impact on host cells and threatens the productivity of bacterial bioprocesses in biotechnology facilities, which can lead to widespread fermentation failure and immeasurable economic loss. Host-controlled defense systems can be developed into effective genetic engineering solutions to address bacteriophage contamination in industrial-scale fermentation; however, most of the resistance mechanisms only narrowly restrict phages and their effect on phage contamination will be limited.
There have been attempts to develop diverse abilities/systems for environmental adaptation or antiviral defense. The team’s collaborative efforts developed a new type II single-stranded DNA phosphorothioation (Ssp) defense system derived from E. coli 3234/A, which can be used in multiple industrial E. coli strains (e.g., E. coli K-12, B and W) to provide broad protection against various types of dsDNA coliphages. Furthermore, they developed a systematic genome engineering strategy involving the simultaneous genomic integration of the Ssp defense module and mutations in components that are essential to the phage life cycle. This strategy can be used to transform E. coli hosts that are highly susceptible to phage attack into strains with powerful restriction effects on the tested bacteriophages. This endows hosts with strong resistance against a wide spectrum of phage infections without affecting bacterial growth and normal physiological function. More importantly, the resulting engineered phage-resistant strains maintained the capabilities of producing the desired chemicals and recombinant proteins even under high levels of phage cocktail challenge, which provides crucial protection against phage attacks.
This is a major step forward, as it provides a systematic solution for engineering phage-resistant bacterial strains, especially industrial bioproduction strains, to protect cells from a wide range of bacteriophages. Considering the functionality of this engineering strategy with diverse E. coli strains, the strategy reported in this study can be widely extended to other bacterial species and industrial applications, which will be of great interest to researchers in academia and industry alike.
Fig. A schematic model of the systematic strategy for engineering phage-sensitive industrial E. coli strains into strains with broad antiphage activities. Through the simultaneous genomic integration of a DNA phosphorothioation-based Ssp defense module and mutations of components essential for the phage life cycle, the engineered E. coli strains show strong resistance against diverse phages tested and maintain the capabilities of producing example recombinant proteins, even under high levels of phage cocktail challenge.
2022.08.23 View 15172 -
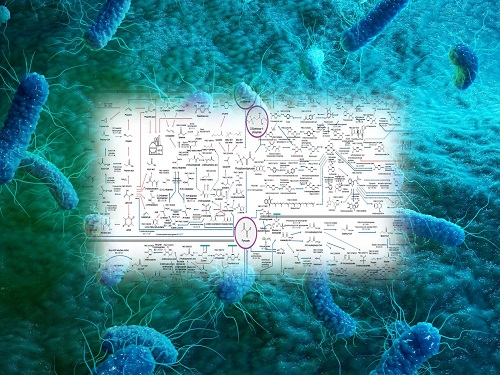 Interactive Map of Metabolical Synthesis of Chemicals
An interactive map that compiled the chemicals produced by biological, chemical and combined reactions has been distributed on the web
- A team led by Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering, organized and distributed an all-inclusive listing of chemical substances that can be synthesized using microorganisms
- It is expected to be used by researchers around the world as it enables easy assessment of the synthetic pathway through the web.
A research team comprised of Woo Dae Jang, Gi Bae Kim, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST reported an interactive metabolic map of bio-based chemicals. Their research paper “An interactive metabolic map of bio-based chemicals” was published online in Trends in Biotechnology on August 10, 2022.
As a response to rapid climate change and environmental pollution, research on the production of petrochemical products using microorganisms is receiving attention as a sustainable alternative to existing methods of productions. In order to synthesize various chemical substances, materials, and fuel using microorganisms, it is necessary to first construct the biosynthetic pathway toward desired product by exploration and discovery and introduce them into microorganisms. In addition, in order to efficiently synthesize various chemical substances, it is sometimes necessary to employ chemical methods along with bioengineering methods using microorganisms at the same time. For the production of non-native chemicals, novel pathways are designed by recruiting enzymes from heterologous sources or employing enzymes designed though rational engineering, directed evolution, or ab initio design.
The research team had completed a map of chemicals which compiled all available pathways of biological and/or chemical reactions that lead to the production of various bio-based chemicals back in 2019 and published the map in Nature Catalysis. The map was distributed in the form of a poster to industries and academia so that the synthesis paths of bio-based chemicals could be checked at a glance.
The research team has expanded the bio-based chemicals map this time in the form of an interactive map on the web so that anyone with internet access can quickly explore efficient paths to synthesize desired products. The web-based map provides interactive visual tools to allow interactive visualization, exploration, and analysis of complex networks of biological and/or chemical reactions toward the desired products. In addition, the reported paper also discusses the production of natural compounds that are used for diverse purposes such as food and medicine, which will help designing novel pathways through similar approaches or by exploiting the promiscuity of enzymes described in the map. The published bio-based chemicals map is also available at http://systemsbiotech.co.kr.
The co-first authors, Dr. Woo Dae Jang and Ph.D. student Gi Bae Kim, said, “We conducted this study to address the demand for updating the previously distributed chemicals map and enhancing its versatility.” “The map is expected to be utilized in a variety of research and in efforts to set strategies and prospects for chemical production incorporating bio and chemical methods that are detailed in the map.”
Distinguished Professor Sang Yup Lee said, “The interactive bio-based chemicals map is expected to help design and optimization of the metabolic pathways for the biosynthesis of target chemicals together with the strategies of chemical conversions, serving as a blueprint for developing further ideas on the production of desired chemicals through biological and/or chemical reactions.”
The interactive metabolic map of bio-based chemicals.
2022.08.11 View 16464
Interactive Map of Metabolical Synthesis of Chemicals
An interactive map that compiled the chemicals produced by biological, chemical and combined reactions has been distributed on the web
- A team led by Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering, organized and distributed an all-inclusive listing of chemical substances that can be synthesized using microorganisms
- It is expected to be used by researchers around the world as it enables easy assessment of the synthetic pathway through the web.
A research team comprised of Woo Dae Jang, Gi Bae Kim, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST reported an interactive metabolic map of bio-based chemicals. Their research paper “An interactive metabolic map of bio-based chemicals” was published online in Trends in Biotechnology on August 10, 2022.
As a response to rapid climate change and environmental pollution, research on the production of petrochemical products using microorganisms is receiving attention as a sustainable alternative to existing methods of productions. In order to synthesize various chemical substances, materials, and fuel using microorganisms, it is necessary to first construct the biosynthetic pathway toward desired product by exploration and discovery and introduce them into microorganisms. In addition, in order to efficiently synthesize various chemical substances, it is sometimes necessary to employ chemical methods along with bioengineering methods using microorganisms at the same time. For the production of non-native chemicals, novel pathways are designed by recruiting enzymes from heterologous sources or employing enzymes designed though rational engineering, directed evolution, or ab initio design.
The research team had completed a map of chemicals which compiled all available pathways of biological and/or chemical reactions that lead to the production of various bio-based chemicals back in 2019 and published the map in Nature Catalysis. The map was distributed in the form of a poster to industries and academia so that the synthesis paths of bio-based chemicals could be checked at a glance.
The research team has expanded the bio-based chemicals map this time in the form of an interactive map on the web so that anyone with internet access can quickly explore efficient paths to synthesize desired products. The web-based map provides interactive visual tools to allow interactive visualization, exploration, and analysis of complex networks of biological and/or chemical reactions toward the desired products. In addition, the reported paper also discusses the production of natural compounds that are used for diverse purposes such as food and medicine, which will help designing novel pathways through similar approaches or by exploiting the promiscuity of enzymes described in the map. The published bio-based chemicals map is also available at http://systemsbiotech.co.kr.
The co-first authors, Dr. Woo Dae Jang and Ph.D. student Gi Bae Kim, said, “We conducted this study to address the demand for updating the previously distributed chemicals map and enhancing its versatility.” “The map is expected to be utilized in a variety of research and in efforts to set strategies and prospects for chemical production incorporating bio and chemical methods that are detailed in the map.”
Distinguished Professor Sang Yup Lee said, “The interactive bio-based chemicals map is expected to help design and optimization of the metabolic pathways for the biosynthesis of target chemicals together with the strategies of chemical conversions, serving as a blueprint for developing further ideas on the production of desired chemicals through biological and/or chemical reactions.”
The interactive metabolic map of bio-based chemicals.
2022.08.11 View 16464 -
 Shaping the AI Semiconductor Ecosystem
- As the marriage of AI and semiconductor being highlighted as the strategic technology of national enthusiasm, KAIST's achievements in the related fields accumulated through top-class education and research capabilities that surpass that of peer universities around the world are standing far apart from the rest of the pack.
As Artificial Intelligence Semiconductor, or a system of semiconductors designed for specifically for highly complicated computation need for AI to conduct its learning and deducing calculations, (hereafter AI semiconductors) stand out as a national strategic technology, the related achievements of KAIST, headed by President Kwang Hyung Lee, are also attracting attention. The Ministry of Science, ICT and Future Planning (MSIT) of Korea initiated a program to support the advancement of AI semiconductor last year with the goal of occupying 20% of the global AI semiconductor market by 2030. This year, through industry-university-research discussions, the Ministry expanded to the program with the addition of 1.2 trillion won of investment over five years through 'Support Plan for AI Semiconductor Industry Promotion'. Accordingly, major universities began putting together programs devised to train students to develop expertise in AI semiconductors.
KAIST has accumulated top-notch educational and research capabilities in the two core fields of AI semiconductor - Semiconductor and Artificial Intelligence. Notably, in the field of semiconductors, the International Solid-State Circuit Conference (ISSCC) is the world's most prestigious conference about designing of semiconductor integrated circuit. Established in 1954, with more than 60% of the participants coming from companies including Samsung, Qualcomm, TSMC, and Intel, the conference naturally focuses on practical value of the studies from the industrial point-of-view, earning the nickname the ‘Semiconductor Design Olympics’. At such conference of legacy and influence, KAIST kept its presence widely visible over other participating universities, leading in terms of the number of accepted papers over world-class schools such as Massachusetts Institute of Technology (MIT) and Stanford for the past 17 years.
Number of papers published at the InternationalSolid-State Circuit Conference (ISSCC) in 2022 sorted by nations and by institutions
Number of papers by universities presented at the International Solid-State Circuit Conference (ISCCC) in 2006~2022
In terms of the number of papers accepted at the ISSCC, KAIST ranked among top two universities each year since 2006. Looking at the average number of accepted papers over the past 17 years, KAIST stands out as an unparalleled leader. The average number of KAIST papers adopted during the period of 17 years from 2006 through 2022, was 8.4, which is almost double of that of competitors like MIT (4.6) and UCLA (3.6). In Korea, it maintains the second place overall after Samsung, the undisputed number one in the semiconductor design field. Also, this year, KAIST was ranked first among universities participating at the Symposium on VLSI Technology and Circuits, an academic conference in the field of integrated circuits that rivals the ISSCC.
Number of papers adopted by the Symposium on VLSI Technology and Circuits in 2022 submitted from the universities
With KAIST researchers working and presenting new technologies at the frontiers of all key areas of the semiconductor industry, the quality of KAIST research is also maintained at the highest level. Professor Myoungsoo Jung's research team in the School of Electrical Engineering is actively working to develop heterogeneous computing environment with high energy efficiency in response to the industry's demand for high performance at low power. In the field of materials, a research team led by Professor Byong-Guk Park of the Department of Materials Science and Engineering developed the Spin Orbit Torque (SOT)-based Magnetic RAM (MRAM) memory that operates at least 10 times faster than conventional memories to suggest a way to overcome the limitations of the existing 'von Neumann structure'.
As such, while providing solutions to major challenges in the current semiconductor industry, the development of new technologies necessary to preoccupy new fields in the semiconductor industry are also very actively pursued. In the field of Quantum Computing, which is attracting attention as next-generation computing technology needed in order to take the lead in the fields of cryptography and nonlinear computation, Professor Sanghyeon Kim's research team in the School of Electrical Engineering presented the world's first 3D integrated quantum computing system at 2021 VLSI Symposium. In Neuromorphic Computing, which is expected to bring remarkable advancements in the field of artificial intelligence by utilizing the principles of the neurology, the research team of Professor Shinhyun Choi of School of Electrical Engineering is developing a next-generation memristor that mimics neurons.
The number of papers by the International Conference on Machine Learning (ICML) and the Conference on Neural Information Processing Systems (NeurIPS), two of the world’s most prestigious academic societies in the field of artificial intelligence (KAIST 6th in the world, 1st in Asia, in 2020)
The field of artificial intelligence has also grown rapidly. Based on the number of papers from the International Conference on Machine Learning (ICML) and the Conference on Neural Information Processing Systems (NeurIPS), two of the world's most prestigious conferences in the field of artificial intelligence, KAIST ranked 6th in the world in 2020 and 1st in Asia. Since 2012, KAIST's ranking steadily inclined from 37th to 6th, climbing 31 steps over the period of eight years. In 2021, 129 papers, or about 40%, of Korean papers published at 11 top artificial intelligence conferences were presented by KAIST. Thanks to KAIST's efforts, in 2021, Korea ranked sixth after the United States, China, United Kingdom, Canada, and Germany in terms of the number of papers published by global AI academic societies.
Number of papers from Korea (and by KAIST) published at 11 top conferences in the field of artificial intelligence in 2021
In terms of content, KAIST's AI research is also at the forefront. Professor Hoi-Jun Yoo's research team in the School of Electrical Engineering compensated for the shortcomings of the “edge networks” by implementing artificial intelligence real-time learning networks on mobile devices. In order to materialize artificial intelligence, data accumulation and a huge amount of computation is required. For this, a high-performance server takes care of massive computation, and for the user terminals, the “edge network” that collects data and performs simple computations are used. Professor Yoo's research greatly increased AI’s processing speed and performance by allotting the learning task to the user terminal as well.
In June, a research team led by Professor Min-Soo Kim of the School of Computing presented a solution that is essential for processing super-scale artificial intelligence models. The super-scale machine learning system developed by the research team is expected to achieve speeds up to 8.8 times faster than Google's Tensorflow or IBM's System DS, which are mainly used in the industry.
KAIST is also making remarkable achievements in the field of AI semiconductors. In 2020, Professor Minsoo Rhu's research team in the School of Electrical Engineering succeeded in developing the world's first AI semiconductor optimized for AI recommendation systems. Due to the nature of the AI recommendation system having to handle vast amounts of contents and user information, it quickly meets its limitation because of the information bottleneck when the process is operated through a general-purpose artificial intelligence system. Professor Minsoo Rhu's team developed a semiconductor that can achieve a speed that is 21 times faster than existing systems using the 'Processing-In-Memory (PIM)' technology. PIM is a technology that improves efficiency by performing the calculations in 'RAM', or random-access memory, which is usually only used to store data temporarily just before they are processed. When PIM technology is put out on the market, it is expected that fortify competitiveness of Korean companies in the AI semiconductor market drastically, as they already hold great strength in the memory area.
KAIST does not plan to be complacent with its achievements, but is making various plans to further the distance from the competitors catching on in the fields of artificial intelligence, semiconductors, and AI semiconductors. Following the establishment of the first artificial intelligence research center in Korea in 1990, the Kim Jaechul AI Graduate School was opened in 2019 to sustain the supply chain of the experts in the field. In 2020, Artificial Intelligence Semiconductor System Research Center was launched to conduct convergent research on AI and semiconductors, which was followed by the establishment of the AI Institutes to promote “AI+X” research efforts.
Based on the internal capabilities accumulated through these efforts, KAIST is also making efforts to train human resources needed in these areas. KAIST established joint research centers with companies such as Naver, while collaborating with local governments such as Hwaseong City to simultaneously nurture professional manpower. Back in 2021, KAIST signed an agreement to establish the Semiconductor System Engineering Department with Samsung Electronics and are preparing a new semiconductor specialist training program. The newly established Department of Semiconductor System Engineering will select around 100 new students every year from 2023 and provide special scholarships to all students so that they can develop their professional skills. In addition, through close cooperation with the industry, they will receive special support which includes field trips and internships at Samsung Electronics, and joint workshops and on-site training.
KAIST has made a significant contribution to the growth of the Korean semiconductor industry ecosystem, producing 25% of doctoral workers in the domestic semiconductor field and 20% of CEOs of mid-sized and venture companies with doctoral degrees. With the dawn coming up on the AI semiconductor ecosystem, whether KAIST will reprise the pivotal role seems to be the crucial point of business.
2022.08.05 View 13439
Shaping the AI Semiconductor Ecosystem
- As the marriage of AI and semiconductor being highlighted as the strategic technology of national enthusiasm, KAIST's achievements in the related fields accumulated through top-class education and research capabilities that surpass that of peer universities around the world are standing far apart from the rest of the pack.
As Artificial Intelligence Semiconductor, or a system of semiconductors designed for specifically for highly complicated computation need for AI to conduct its learning and deducing calculations, (hereafter AI semiconductors) stand out as a national strategic technology, the related achievements of KAIST, headed by President Kwang Hyung Lee, are also attracting attention. The Ministry of Science, ICT and Future Planning (MSIT) of Korea initiated a program to support the advancement of AI semiconductor last year with the goal of occupying 20% of the global AI semiconductor market by 2030. This year, through industry-university-research discussions, the Ministry expanded to the program with the addition of 1.2 trillion won of investment over five years through 'Support Plan for AI Semiconductor Industry Promotion'. Accordingly, major universities began putting together programs devised to train students to develop expertise in AI semiconductors.
KAIST has accumulated top-notch educational and research capabilities in the two core fields of AI semiconductor - Semiconductor and Artificial Intelligence. Notably, in the field of semiconductors, the International Solid-State Circuit Conference (ISSCC) is the world's most prestigious conference about designing of semiconductor integrated circuit. Established in 1954, with more than 60% of the participants coming from companies including Samsung, Qualcomm, TSMC, and Intel, the conference naturally focuses on practical value of the studies from the industrial point-of-view, earning the nickname the ‘Semiconductor Design Olympics’. At such conference of legacy and influence, KAIST kept its presence widely visible over other participating universities, leading in terms of the number of accepted papers over world-class schools such as Massachusetts Institute of Technology (MIT) and Stanford for the past 17 years.
Number of papers published at the InternationalSolid-State Circuit Conference (ISSCC) in 2022 sorted by nations and by institutions
Number of papers by universities presented at the International Solid-State Circuit Conference (ISCCC) in 2006~2022
In terms of the number of papers accepted at the ISSCC, KAIST ranked among top two universities each year since 2006. Looking at the average number of accepted papers over the past 17 years, KAIST stands out as an unparalleled leader. The average number of KAIST papers adopted during the period of 17 years from 2006 through 2022, was 8.4, which is almost double of that of competitors like MIT (4.6) and UCLA (3.6). In Korea, it maintains the second place overall after Samsung, the undisputed number one in the semiconductor design field. Also, this year, KAIST was ranked first among universities participating at the Symposium on VLSI Technology and Circuits, an academic conference in the field of integrated circuits that rivals the ISSCC.
Number of papers adopted by the Symposium on VLSI Technology and Circuits in 2022 submitted from the universities
With KAIST researchers working and presenting new technologies at the frontiers of all key areas of the semiconductor industry, the quality of KAIST research is also maintained at the highest level. Professor Myoungsoo Jung's research team in the School of Electrical Engineering is actively working to develop heterogeneous computing environment with high energy efficiency in response to the industry's demand for high performance at low power. In the field of materials, a research team led by Professor Byong-Guk Park of the Department of Materials Science and Engineering developed the Spin Orbit Torque (SOT)-based Magnetic RAM (MRAM) memory that operates at least 10 times faster than conventional memories to suggest a way to overcome the limitations of the existing 'von Neumann structure'.
As such, while providing solutions to major challenges in the current semiconductor industry, the development of new technologies necessary to preoccupy new fields in the semiconductor industry are also very actively pursued. In the field of Quantum Computing, which is attracting attention as next-generation computing technology needed in order to take the lead in the fields of cryptography and nonlinear computation, Professor Sanghyeon Kim's research team in the School of Electrical Engineering presented the world's first 3D integrated quantum computing system at 2021 VLSI Symposium. In Neuromorphic Computing, which is expected to bring remarkable advancements in the field of artificial intelligence by utilizing the principles of the neurology, the research team of Professor Shinhyun Choi of School of Electrical Engineering is developing a next-generation memristor that mimics neurons.
The number of papers by the International Conference on Machine Learning (ICML) and the Conference on Neural Information Processing Systems (NeurIPS), two of the world’s most prestigious academic societies in the field of artificial intelligence (KAIST 6th in the world, 1st in Asia, in 2020)
The field of artificial intelligence has also grown rapidly. Based on the number of papers from the International Conference on Machine Learning (ICML) and the Conference on Neural Information Processing Systems (NeurIPS), two of the world's most prestigious conferences in the field of artificial intelligence, KAIST ranked 6th in the world in 2020 and 1st in Asia. Since 2012, KAIST's ranking steadily inclined from 37th to 6th, climbing 31 steps over the period of eight years. In 2021, 129 papers, or about 40%, of Korean papers published at 11 top artificial intelligence conferences were presented by KAIST. Thanks to KAIST's efforts, in 2021, Korea ranked sixth after the United States, China, United Kingdom, Canada, and Germany in terms of the number of papers published by global AI academic societies.
Number of papers from Korea (and by KAIST) published at 11 top conferences in the field of artificial intelligence in 2021
In terms of content, KAIST's AI research is also at the forefront. Professor Hoi-Jun Yoo's research team in the School of Electrical Engineering compensated for the shortcomings of the “edge networks” by implementing artificial intelligence real-time learning networks on mobile devices. In order to materialize artificial intelligence, data accumulation and a huge amount of computation is required. For this, a high-performance server takes care of massive computation, and for the user terminals, the “edge network” that collects data and performs simple computations are used. Professor Yoo's research greatly increased AI’s processing speed and performance by allotting the learning task to the user terminal as well.
In June, a research team led by Professor Min-Soo Kim of the School of Computing presented a solution that is essential for processing super-scale artificial intelligence models. The super-scale machine learning system developed by the research team is expected to achieve speeds up to 8.8 times faster than Google's Tensorflow or IBM's System DS, which are mainly used in the industry.
KAIST is also making remarkable achievements in the field of AI semiconductors. In 2020, Professor Minsoo Rhu's research team in the School of Electrical Engineering succeeded in developing the world's first AI semiconductor optimized for AI recommendation systems. Due to the nature of the AI recommendation system having to handle vast amounts of contents and user information, it quickly meets its limitation because of the information bottleneck when the process is operated through a general-purpose artificial intelligence system. Professor Minsoo Rhu's team developed a semiconductor that can achieve a speed that is 21 times faster than existing systems using the 'Processing-In-Memory (PIM)' technology. PIM is a technology that improves efficiency by performing the calculations in 'RAM', or random-access memory, which is usually only used to store data temporarily just before they are processed. When PIM technology is put out on the market, it is expected that fortify competitiveness of Korean companies in the AI semiconductor market drastically, as they already hold great strength in the memory area.
KAIST does not plan to be complacent with its achievements, but is making various plans to further the distance from the competitors catching on in the fields of artificial intelligence, semiconductors, and AI semiconductors. Following the establishment of the first artificial intelligence research center in Korea in 1990, the Kim Jaechul AI Graduate School was opened in 2019 to sustain the supply chain of the experts in the field. In 2020, Artificial Intelligence Semiconductor System Research Center was launched to conduct convergent research on AI and semiconductors, which was followed by the establishment of the AI Institutes to promote “AI+X” research efforts.
Based on the internal capabilities accumulated through these efforts, KAIST is also making efforts to train human resources needed in these areas. KAIST established joint research centers with companies such as Naver, while collaborating with local governments such as Hwaseong City to simultaneously nurture professional manpower. Back in 2021, KAIST signed an agreement to establish the Semiconductor System Engineering Department with Samsung Electronics and are preparing a new semiconductor specialist training program. The newly established Department of Semiconductor System Engineering will select around 100 new students every year from 2023 and provide special scholarships to all students so that they can develop their professional skills. In addition, through close cooperation with the industry, they will receive special support which includes field trips and internships at Samsung Electronics, and joint workshops and on-site training.
KAIST has made a significant contribution to the growth of the Korean semiconductor industry ecosystem, producing 25% of doctoral workers in the domestic semiconductor field and 20% of CEOs of mid-sized and venture companies with doctoral degrees. With the dawn coming up on the AI semiconductor ecosystem, whether KAIST will reprise the pivotal role seems to be the crucial point of business.
2022.08.05 View 13439 -
 KAIST Honors BMW and Hyundai with the 2022 Future Mobility of the Year Award
BMW ‘iVision Circular’, Commercial Vehicle-Hyundai Motors ‘Trailer Drone’ selected as winners of the international awards for concept cars established by KAIST Cho Chun Shik Graduate School of Mobility to honor car makers that strive to present new visions in the field of eco-friendly design of automobiles and unmanned logistics.
KAIST (President Kwang Hyung Lee) hosted the “2022 Future Mobility of the Year (FMOTY) Awards” at the Convention Hall of the BEXCO International Motor Show at Busan in the afternoon of the 14th.
The Future Mobility of the Year Awards is an award ceremony that selects a model that showcases useful transportation technology and innovative service concepts for the future society among the set of concept cars exhibited at the motor show.
As a one-of-a-kind international concept car awards established by KAIST's Cho Chun Shik Graduate School of Mobility (Headed by Professor Jang In-Gwon), the auto journalists from 11 countries were invited to be the jurors to select the winner. With the inaugural awards ceremony held in 2019, over the past three years, automakers from around the globe, including internationally renowned automakers, such as, Volvo/Toyota (2019), Honda/Hyundai (2020), and Renault (2021), even a new start-up car manufacturer like Canoo, the winner of last year’s award for commercial vehicles, were honored for their award-winning works.
At this year’s awards ceremony, the 4th of its kind, BMW's “iVision Circular” and Hyundai's “'Trailer Drone” were selected as the best concept cars of the year, the former from the Private Mobility category and the latter from the Public & Commercial Vehicles category.
The jury consisting of 16 domestic and foreign auto journalists, including BBC Top Gear's Paul Horrell and Car Magazine’s Georg Kacher, evaluated 53 concept car contestants that made their entry last year. The jurors’ general comment was that while the trend of the global automobile market flowing fast towards electric vehicles, this year's award-winning works presented a new vision in the field of eco-friendly design and unmanned logistics.
Private Mobility Categry Winner: BMW iVision Circular
BMW's 'iVision Circular', the winner of the Private Mobility category, is an eco-friendly compact car in which all parts of the vehicle are designed with recycled and/or natural materials. It has received favorable reviews for its in-depth implementation of the concept of a futuristic eco-friendly car by manufacturing the tires from natural rubber and adopting a design that made recycling of its parts very easily when the car is to be disposed of.
Public & Commercial Vehicles Categry Winner: Hyundai Trailer Drone
Hyundai Motor Company’s “Trailer Drone”, the winner of the Public & Commercial Vehicles category, is an eco-friendly autonomous driving truck that can transport large-scale logistics from a port to a destination without a human driver while two unmanned vehicles push and drag a trailer. The concept car won supports from a large number of judges for the blueprint it presented for a groundbreaking logistics service that applied both eco-friendly hydrogen fuel cell and fully autonomous driving technology.
Jurors from overseas congratulated the development team of BMW and Hyundai Motor Company via a video message for providing a new direction for the global automobile industry as it strives to transform in line with the changes in the post-pandemic era.
Professor Bo-won Kim, the Vice President for Planning and Budget of KAIST, who presented the awards, said, “It is time for the K-Mobility wave to sweep over the global mobility industry.” “KAIST will lead in the various fields of mobility technologies to support global automakers,” he added.
Splitting the center are KAIST Vice President Bo-Won Kim on the right, and Seong-Kwon Lee,
the Deputy Mayor of the City of Busan on the left. To Kim's left is the Senior VP of BMW Asia-Pacific, Eastern Europe, Middle East, Africa, Jean-Philippe Parain, and to Lee's Right is Sangyup Lee,
the Head of Hyundai Motor Design Center and the Executive VP of Hyundai Motors.
At the ceremony, along with KAIST officials, including Vice President Bo-Won Kim and Professor In-Gwon Jang, the Head of Cho Chun Shik Graduate School of Mobility, are the Deputy Mayor Seong-Kwon Lee of the City of Busan and the figures from the automobile industry, including Jean-Philippe Parain, the Senior Vice President of BMW Asia-Pacific, Eastern Europe, Middle East, Africa, who is visiting Korea to receive the '2022 Future Mobility' award, and Sangyup Lee, the Head of Hyundai Motor Design Center and the Executive Vice President of Hyundai Motor Company, were in the attendance.
More information about the awards ceremony and winning works are available at the official website of this year's Future Mobility Awards (www.fmoty.org).
Profile:In-Gwon Jang, Ph.D.Presidentthe Organizing Committeethe Future Mobility of the Year Awardshttp://www.fmoty.org/
Head ProfessorKAIST Cho Chun Shik Graduate School of Mobilityhttps://gt.kaist.ac.kr
2022.07.14 View 16244
KAIST Honors BMW and Hyundai with the 2022 Future Mobility of the Year Award
BMW ‘iVision Circular’, Commercial Vehicle-Hyundai Motors ‘Trailer Drone’ selected as winners of the international awards for concept cars established by KAIST Cho Chun Shik Graduate School of Mobility to honor car makers that strive to present new visions in the field of eco-friendly design of automobiles and unmanned logistics.
KAIST (President Kwang Hyung Lee) hosted the “2022 Future Mobility of the Year (FMOTY) Awards” at the Convention Hall of the BEXCO International Motor Show at Busan in the afternoon of the 14th.
The Future Mobility of the Year Awards is an award ceremony that selects a model that showcases useful transportation technology and innovative service concepts for the future society among the set of concept cars exhibited at the motor show.
As a one-of-a-kind international concept car awards established by KAIST's Cho Chun Shik Graduate School of Mobility (Headed by Professor Jang In-Gwon), the auto journalists from 11 countries were invited to be the jurors to select the winner. With the inaugural awards ceremony held in 2019, over the past three years, automakers from around the globe, including internationally renowned automakers, such as, Volvo/Toyota (2019), Honda/Hyundai (2020), and Renault (2021), even a new start-up car manufacturer like Canoo, the winner of last year’s award for commercial vehicles, were honored for their award-winning works.
At this year’s awards ceremony, the 4th of its kind, BMW's “iVision Circular” and Hyundai's “'Trailer Drone” were selected as the best concept cars of the year, the former from the Private Mobility category and the latter from the Public & Commercial Vehicles category.
The jury consisting of 16 domestic and foreign auto journalists, including BBC Top Gear's Paul Horrell and Car Magazine’s Georg Kacher, evaluated 53 concept car contestants that made their entry last year. The jurors’ general comment was that while the trend of the global automobile market flowing fast towards electric vehicles, this year's award-winning works presented a new vision in the field of eco-friendly design and unmanned logistics.
Private Mobility Categry Winner: BMW iVision Circular
BMW's 'iVision Circular', the winner of the Private Mobility category, is an eco-friendly compact car in which all parts of the vehicle are designed with recycled and/or natural materials. It has received favorable reviews for its in-depth implementation of the concept of a futuristic eco-friendly car by manufacturing the tires from natural rubber and adopting a design that made recycling of its parts very easily when the car is to be disposed of.
Public & Commercial Vehicles Categry Winner: Hyundai Trailer Drone
Hyundai Motor Company’s “Trailer Drone”, the winner of the Public & Commercial Vehicles category, is an eco-friendly autonomous driving truck that can transport large-scale logistics from a port to a destination without a human driver while two unmanned vehicles push and drag a trailer. The concept car won supports from a large number of judges for the blueprint it presented for a groundbreaking logistics service that applied both eco-friendly hydrogen fuel cell and fully autonomous driving technology.
Jurors from overseas congratulated the development team of BMW and Hyundai Motor Company via a video message for providing a new direction for the global automobile industry as it strives to transform in line with the changes in the post-pandemic era.
Professor Bo-won Kim, the Vice President for Planning and Budget of KAIST, who presented the awards, said, “It is time for the K-Mobility wave to sweep over the global mobility industry.” “KAIST will lead in the various fields of mobility technologies to support global automakers,” he added.
Splitting the center are KAIST Vice President Bo-Won Kim on the right, and Seong-Kwon Lee,
the Deputy Mayor of the City of Busan on the left. To Kim's left is the Senior VP of BMW Asia-Pacific, Eastern Europe, Middle East, Africa, Jean-Philippe Parain, and to Lee's Right is Sangyup Lee,
the Head of Hyundai Motor Design Center and the Executive VP of Hyundai Motors.
At the ceremony, along with KAIST officials, including Vice President Bo-Won Kim and Professor In-Gwon Jang, the Head of Cho Chun Shik Graduate School of Mobility, are the Deputy Mayor Seong-Kwon Lee of the City of Busan and the figures from the automobile industry, including Jean-Philippe Parain, the Senior Vice President of BMW Asia-Pacific, Eastern Europe, Middle East, Africa, who is visiting Korea to receive the '2022 Future Mobility' award, and Sangyup Lee, the Head of Hyundai Motor Design Center and the Executive Vice President of Hyundai Motor Company, were in the attendance.
More information about the awards ceremony and winning works are available at the official website of this year's Future Mobility Awards (www.fmoty.org).
Profile:In-Gwon Jang, Ph.D.Presidentthe Organizing Committeethe Future Mobility of the Year Awardshttp://www.fmoty.org/
Head ProfessorKAIST Cho Chun Shik Graduate School of Mobilityhttps://gt.kaist.ac.kr
2022.07.14 View 16244 -
 An AI-based, Indoor/Outdoor-Integrated (IOI) GPS System to Bring Seismic Waves in the Terrains of Positioning Technology
KAIST breaks new grounds in positioning technology with an AI-integrated GPS board that works both indoors and out
KAIST (President Kwang Hyung Lee) announced on the 8th that Professor Dong-Soo Han's research team (Intelligent Service Integration Lab) from the School of Computing has developed a GPS system that works both indoors and outdoors with quality precision regardless of the environment.
This Indoor/Outdoor-Integrated GPS System, or IOI GPS System, for short, uses the GPS signals outdoors and estimates locations indoors using signals from multiple sources like an inertial sensor, pressure sensors, geomagnetic sensors, and light sensors. To this end, the research team developed techniques to detect environmental changes such as entering a building, and methods to detect entrances, ground floors, stairs, elevators and levels of buildings by utilizing artificial intelligence techniques. Various landmark detecting techniques were also incorporated with pedestrian dead reckoning (PDR), a navigation tool for pedestrians, to devise the so-called “Sensor-Fusion Positioning Algorithm”.
To date, it was common to estimate locations based on wireless LAN signals or base station signals in a space where the GPS signal could not reach. However, the IOI GPS enables positioning even in buildings without signals nor indoor maps.
The algorithm developed by the research team can provide accurate floor information within a building where even big tech companies like Google and Apple's positioning services do not provide. Unlike other positioning methods that rely on visual data, geomagnetic positioning techniques, or wireless LAN, this system also has the advantage of not requiring any prior preparation. In other words, the foundation to enable the usage of a universal GPS system that works both indoors and outdoors anywhere in the world is now ready.
The research team also produced a circuit board for the purpose of operating the IOI GPS System, mounted with chips to receive and process GPS, Wi-Fi, and Bluetooth signals, along with an inertial sensor, a barometer, a magnetometer, and a light sensor. The sensor-fusion positioning algorithm the lab has developed is also incorporated in the board.
When the accuracy of the IOI GPS board was tested in the N1 building of KAIST’s main campus in Daejeon, it achieved an accuracy of about 95% in floor estimation and an accuracy of about 3 to 6 meters in distance estimation. As for the indoor/outdoor transition, the navigational mode change was completed in about 0.3 seconds. When it was combined with the PDR technique, the estimation accuracy improved further down to a scope of one meter.
The research team is now working on assembling a tag with a built-in positioning board and applying it to location-based docent services for visitors at museums, science centers, and art galleries. The IOI GPS tag can be used for the purpose of tracking children and/or the elderly, and it can also be used to locate people or rescue workers lost in disaster-ridden or hazardous sites. On a different note, the sensor-fusion positioning algorithm and positioning board for vehicles are also under development for the tracking of vehicles entering indoor areas like underground parking lots.
When the IOI GPS board for vehicles is manufactured, the research team will work to collaborate with car manufacturers and car rental companies, and will also develop a sensor-fusion positioning algorithm for smartphones. Telecommunication companies seeking to diversify their programs in the field of location-based services will also be interested in the use the IOI GPS.
Professor Dong-Soo Han of the School of Computing, who leads the research team, said, “This is the first time to develop an indoor/outdoor integrated GPS system that can pinpoint locations in a building where there is no wireless signal or an indoor map, and there are an infinite number of areas it can be applied to. When the integration with the Korea Augmentation Satellite System (KASS) and the Korean GPS (KPS) System that began this year, is finally completed, Korea can become the leader in the field of GPS both indoors and outdoors, and we also have plans to manufacture semi-conductor chips for the IOI GPS System to keep the tech-gap between Korea and the followers.”
He added, "The guidance services at science centers, museums, and art galleries that uses IOI GPS tags can provide a set of data that would be very helpful for analyzing the visitors’ viewing traces. It is an essential piece of information required when the time comes to decide when to organize the next exhibit. We will be working on having it applied to the National Science Museum, first.”
The projects to develop the IOI GPS system and the trace analysis system for science centers were supported through Science, Culture, Exhibits and Services Capability Enhancement Program of the Ministry of Science and ICT.
Profile: Dong-Soo Han, Ph.D.Professorddsshhan@kaist.ac.krhttp://isilab.kaist.ac.kr
Intelligent Service Integration Lab.School of Computing
http://kaist.ac.kr/en/
Korea Advanced Institute of Science and Technology (KAIST)Daejeon, Republic of Korea
2022.07.13 View 12735
An AI-based, Indoor/Outdoor-Integrated (IOI) GPS System to Bring Seismic Waves in the Terrains of Positioning Technology
KAIST breaks new grounds in positioning technology with an AI-integrated GPS board that works both indoors and out
KAIST (President Kwang Hyung Lee) announced on the 8th that Professor Dong-Soo Han's research team (Intelligent Service Integration Lab) from the School of Computing has developed a GPS system that works both indoors and outdoors with quality precision regardless of the environment.
This Indoor/Outdoor-Integrated GPS System, or IOI GPS System, for short, uses the GPS signals outdoors and estimates locations indoors using signals from multiple sources like an inertial sensor, pressure sensors, geomagnetic sensors, and light sensors. To this end, the research team developed techniques to detect environmental changes such as entering a building, and methods to detect entrances, ground floors, stairs, elevators and levels of buildings by utilizing artificial intelligence techniques. Various landmark detecting techniques were also incorporated with pedestrian dead reckoning (PDR), a navigation tool for pedestrians, to devise the so-called “Sensor-Fusion Positioning Algorithm”.
To date, it was common to estimate locations based on wireless LAN signals or base station signals in a space where the GPS signal could not reach. However, the IOI GPS enables positioning even in buildings without signals nor indoor maps.
The algorithm developed by the research team can provide accurate floor information within a building where even big tech companies like Google and Apple's positioning services do not provide. Unlike other positioning methods that rely on visual data, geomagnetic positioning techniques, or wireless LAN, this system also has the advantage of not requiring any prior preparation. In other words, the foundation to enable the usage of a universal GPS system that works both indoors and outdoors anywhere in the world is now ready.
The research team also produced a circuit board for the purpose of operating the IOI GPS System, mounted with chips to receive and process GPS, Wi-Fi, and Bluetooth signals, along with an inertial sensor, a barometer, a magnetometer, and a light sensor. The sensor-fusion positioning algorithm the lab has developed is also incorporated in the board.
When the accuracy of the IOI GPS board was tested in the N1 building of KAIST’s main campus in Daejeon, it achieved an accuracy of about 95% in floor estimation and an accuracy of about 3 to 6 meters in distance estimation. As for the indoor/outdoor transition, the navigational mode change was completed in about 0.3 seconds. When it was combined with the PDR technique, the estimation accuracy improved further down to a scope of one meter.
The research team is now working on assembling a tag with a built-in positioning board and applying it to location-based docent services for visitors at museums, science centers, and art galleries. The IOI GPS tag can be used for the purpose of tracking children and/or the elderly, and it can also be used to locate people or rescue workers lost in disaster-ridden or hazardous sites. On a different note, the sensor-fusion positioning algorithm and positioning board for vehicles are also under development for the tracking of vehicles entering indoor areas like underground parking lots.
When the IOI GPS board for vehicles is manufactured, the research team will work to collaborate with car manufacturers and car rental companies, and will also develop a sensor-fusion positioning algorithm for smartphones. Telecommunication companies seeking to diversify their programs in the field of location-based services will also be interested in the use the IOI GPS.
Professor Dong-Soo Han of the School of Computing, who leads the research team, said, “This is the first time to develop an indoor/outdoor integrated GPS system that can pinpoint locations in a building where there is no wireless signal or an indoor map, and there are an infinite number of areas it can be applied to. When the integration with the Korea Augmentation Satellite System (KASS) and the Korean GPS (KPS) System that began this year, is finally completed, Korea can become the leader in the field of GPS both indoors and outdoors, and we also have plans to manufacture semi-conductor chips for the IOI GPS System to keep the tech-gap between Korea and the followers.”
He added, "The guidance services at science centers, museums, and art galleries that uses IOI GPS tags can provide a set of data that would be very helpful for analyzing the visitors’ viewing traces. It is an essential piece of information required when the time comes to decide when to organize the next exhibit. We will be working on having it applied to the National Science Museum, first.”
The projects to develop the IOI GPS system and the trace analysis system for science centers were supported through Science, Culture, Exhibits and Services Capability Enhancement Program of the Ministry of Science and ICT.
Profile: Dong-Soo Han, Ph.D.Professorddsshhan@kaist.ac.krhttp://isilab.kaist.ac.kr
Intelligent Service Integration Lab.School of Computing
http://kaist.ac.kr/en/
Korea Advanced Institute of Science and Technology (KAIST)Daejeon, Republic of Korea
2022.07.13 View 12735 -
 Atomically-Smooth Gold Crystals Help to Compress Light for Nanophotonic Applications
Highly compressed mid-infrared optical waves in a thin dielectric crystal on monocrystalline gold substrate investigated for the first time using a high-resolution scattering-type scanning near-field optical microscope.
KAIST researchers and their collaborators at home and abroad have successfully demonstrated a new platform for guiding the compressed light waves in very thin van der Waals crystals. Their method to guide the mid-infrared light with minimal loss will provide a breakthrough for the practical applications of ultra-thin dielectric crystals in next-generation optoelectronic devices based on strong light-matter interactions at the nanoscale.
Phonon-polaritons are collective oscillations of ions in polar dielectrics coupled to electromagnetic waves of light, whose electromagnetic field is much more compressed compared to the light wavelength. Recently, it was demonstrated that the phonon-polaritons in thin van der Waals crystals can be compressed even further when the material is placed on top of a highly conductive metal. In such a configuration, charges in the polaritonic crystal are “reflected” in the metal, and their coupling with light results in a new type of polariton waves called the image phonon-polaritons. Highly compressed image modes provide strong light-matter interactions, but are very sensitive to the substrate roughness, which hinders their practical application.
Challenged by these limitations, four research groups combined their efforts to develop a unique experimental platform using advanced fabrication and measurement methods. Their findings were published in Science Advances on July 13.
A KAIST research team led by Professor Min Seok Jang from the School of Electrical Engineering used a highly sensitive scanning near-field optical microscope (SNOM) to directly measure the optical fields of the hyperbolic image phonon-polaritons (HIP) propagating in a 63 nm-thick slab of hexagonal boron nitride (h-BN) on a monocrystalline gold substrate, showing the mid-infrared light waves in dielectric crystal compressed by a hundred times.
Professor Jang and a research professor in his group, Sergey Menabde, successfully obtained direct images of HIP waves propagating for many wavelengths, and detected a signal from the ultra-compressed high-order HIP in a regular h-BN crystals for the first time. They showed that the phonon-polaritons in van der Waals crystals can be significantly more compressed without sacrificing their lifetime.
This became possible due to the atomically-smooth surfaces of the home-grown gold crystals used as a substrate for the h-BN. Practically zero surface scattering and extremely small ohmic loss in gold at mid-infrared frequencies provide a low-loss environment for the HIP propagation. The HIP mode probed by the researchers was 2.4 times more compressed and yet exhibited a similar lifetime compared to the phonon-polaritons with a low-loss dielectric substrate, resulting in a twice higher figure of merit in terms of the normalized propagation length.
The ultra-smooth monocrystalline gold flakes used in the experiment were chemically grown by the team of Professor N. Asger Mortensen from the Center for Nano Optics at the University of Southern Denmark.
Mid-infrared spectrum is particularly important for sensing applications since many important organic molecules have absorption lines in the mid-infrared. However, a large number of molecules is required by the conventional detection methods for successful operation, whereas the ultra-compressed phonon-polariton fields can provide strong light-matter interactions at the microscopic level, thus significantly improving the detection limit down to a single molecule. The long lifetime of the HIP on monocrystalline gold will further improve the detection performance.
Furthermore, the study conducted by Professor Jang and the team demonstrated the striking similarity between the HIP and the image graphene plasmons. Both image modes possess significantly more confined electromagnetic field, yet their lifetime remains unaffected by the shorter polariton wavelength. This observation provides a broader perspective on image polaritons in general, and highlights their superiority in terms of the nanolight waveguiding compared to the conventional low-dimensional polaritons in van der Waals crystals on a dielectric substrate.
Professor Jang said, “Our research demonstrated the advantages of image polaritons, and especially the image phonon-polaritons. These optical modes can be used in the future optoelectronic devices where both the low-loss propagation and the strong light-matter interaction are necessary. I hope that our results will pave the way for the realization of more efficient nanophotonic devices such as metasurfaces, optical switches, sensors, and other applications operating at infrared frequencies.”
This research was funded by the Samsung Research Funding & Incubation Center of Samsung Electronics and the National Research Foundation of Korea (NRF). The Korea Institute of Science and Technology, Ministry of Education, Culture, Sports, Science and Technology of Japan, and The Villum Foundation, Denmark, also supported the work.
Figure. Nano-tip is used for the ultra-high-resolution imaging of the image phonon-polaritons in hBN launched by the gold crystal edge.
Publication:
Menabde, S. G., et al. (2022) Near-field probing of image phonon-polaritons in hexagonal boron nitride on gold crystals. Science Advances 8, Article ID: eabn0627. Available online at https://science.org/doi/10.1126/sciadv.abn0627.
Profile:
Min Seok Jang, MS, PhD
Associate Professor
jang.minseok@kaist.ac.kr
http://janglab.org/
Min Seok Jang Research Group
School of Electrical Engineering
http://kaist.ac.kr/en/
Korea Advanced Institute of Science and Technology (KAIST)
Daejeon, Republic of Korea
2022.07.13 View 15174
Atomically-Smooth Gold Crystals Help to Compress Light for Nanophotonic Applications
Highly compressed mid-infrared optical waves in a thin dielectric crystal on monocrystalline gold substrate investigated for the first time using a high-resolution scattering-type scanning near-field optical microscope.
KAIST researchers and their collaborators at home and abroad have successfully demonstrated a new platform for guiding the compressed light waves in very thin van der Waals crystals. Their method to guide the mid-infrared light with minimal loss will provide a breakthrough for the practical applications of ultra-thin dielectric crystals in next-generation optoelectronic devices based on strong light-matter interactions at the nanoscale.
Phonon-polaritons are collective oscillations of ions in polar dielectrics coupled to electromagnetic waves of light, whose electromagnetic field is much more compressed compared to the light wavelength. Recently, it was demonstrated that the phonon-polaritons in thin van der Waals crystals can be compressed even further when the material is placed on top of a highly conductive metal. In such a configuration, charges in the polaritonic crystal are “reflected” in the metal, and their coupling with light results in a new type of polariton waves called the image phonon-polaritons. Highly compressed image modes provide strong light-matter interactions, but are very sensitive to the substrate roughness, which hinders their practical application.
Challenged by these limitations, four research groups combined their efforts to develop a unique experimental platform using advanced fabrication and measurement methods. Their findings were published in Science Advances on July 13.
A KAIST research team led by Professor Min Seok Jang from the School of Electrical Engineering used a highly sensitive scanning near-field optical microscope (SNOM) to directly measure the optical fields of the hyperbolic image phonon-polaritons (HIP) propagating in a 63 nm-thick slab of hexagonal boron nitride (h-BN) on a monocrystalline gold substrate, showing the mid-infrared light waves in dielectric crystal compressed by a hundred times.
Professor Jang and a research professor in his group, Sergey Menabde, successfully obtained direct images of HIP waves propagating for many wavelengths, and detected a signal from the ultra-compressed high-order HIP in a regular h-BN crystals for the first time. They showed that the phonon-polaritons in van der Waals crystals can be significantly more compressed without sacrificing their lifetime.
This became possible due to the atomically-smooth surfaces of the home-grown gold crystals used as a substrate for the h-BN. Practically zero surface scattering and extremely small ohmic loss in gold at mid-infrared frequencies provide a low-loss environment for the HIP propagation. The HIP mode probed by the researchers was 2.4 times more compressed and yet exhibited a similar lifetime compared to the phonon-polaritons with a low-loss dielectric substrate, resulting in a twice higher figure of merit in terms of the normalized propagation length.
The ultra-smooth monocrystalline gold flakes used in the experiment were chemically grown by the team of Professor N. Asger Mortensen from the Center for Nano Optics at the University of Southern Denmark.
Mid-infrared spectrum is particularly important for sensing applications since many important organic molecules have absorption lines in the mid-infrared. However, a large number of molecules is required by the conventional detection methods for successful operation, whereas the ultra-compressed phonon-polariton fields can provide strong light-matter interactions at the microscopic level, thus significantly improving the detection limit down to a single molecule. The long lifetime of the HIP on monocrystalline gold will further improve the detection performance.
Furthermore, the study conducted by Professor Jang and the team demonstrated the striking similarity between the HIP and the image graphene plasmons. Both image modes possess significantly more confined electromagnetic field, yet their lifetime remains unaffected by the shorter polariton wavelength. This observation provides a broader perspective on image polaritons in general, and highlights their superiority in terms of the nanolight waveguiding compared to the conventional low-dimensional polaritons in van der Waals crystals on a dielectric substrate.
Professor Jang said, “Our research demonstrated the advantages of image polaritons, and especially the image phonon-polaritons. These optical modes can be used in the future optoelectronic devices where both the low-loss propagation and the strong light-matter interaction are necessary. I hope that our results will pave the way for the realization of more efficient nanophotonic devices such as metasurfaces, optical switches, sensors, and other applications operating at infrared frequencies.”
This research was funded by the Samsung Research Funding & Incubation Center of Samsung Electronics and the National Research Foundation of Korea (NRF). The Korea Institute of Science and Technology, Ministry of Education, Culture, Sports, Science and Technology of Japan, and The Villum Foundation, Denmark, also supported the work.
Figure. Nano-tip is used for the ultra-high-resolution imaging of the image phonon-polaritons in hBN launched by the gold crystal edge.
Publication:
Menabde, S. G., et al. (2022) Near-field probing of image phonon-polaritons in hexagonal boron nitride on gold crystals. Science Advances 8, Article ID: eabn0627. Available online at https://science.org/doi/10.1126/sciadv.abn0627.
Profile:
Min Seok Jang, MS, PhD
Associate Professor
jang.minseok@kaist.ac.kr
http://janglab.org/
Min Seok Jang Research Group
School of Electrical Engineering
http://kaist.ac.kr/en/
Korea Advanced Institute of Science and Technology (KAIST)
Daejeon, Republic of Korea
2022.07.13 View 15174 -
 The 1st Global Entrepreneurship Summer Camp bridges KAIST and Silicon Valley, US
Twenty KAIST students gave a go at selling their business ideas to investors at Silicon Valley on the “Pitch Day” at 2022 Global Entrepreneurship Summer Camp.
From Tuesday, June 21 to Monday, July 4, 2022, KAIST held the first Global Entrepreneurship Summer Camp (GESC).
The 2022 GESC, which was organized in collaboration with Stanford Technology Ventures Program (STVP), KOTRA Silicon Valley IT Center, and KAIST Alumni at Silicon Valley, was a pilot program that offered opportunities of experiencing and learning about the cases of startup companies in Silicon Valley and a chance to expand businesses to Silicon Valley through networking.
Twenty KAIST students, including pre-startup entrepreneurs and students interested in global entrepreneurship with less than one year of business experience were selected. The first week of the program was organized by Startup KAIST while the second week program was organized by the Center for Global Strategies and Planning (GSP) at KAIST in collaboration with the Stanford Technology Venture Program (STVP), KAIST Alumni at Silicon Valley, and KOTRA at Silicon Valley.
Dr. Mo-Yun Lei Fong, the Executive Director of STVP, said, “The program offered an opportunity for us to realize our vision of empowering aspiring entrepreneurs to become global citizens who create and scale responsible innovation. By collaborating with KAIST and offering entrepreneurial insights to Korean students, we are able to have a positive impact on a global scale.” Mo added, “The program also enabled STVP to build bridges, learn from the students, and refine our culturally relevant curriculum by understanding Korean culture and ideas.”
On the “Pitch Day” on July 1, following a special talk by Dr. Chong-Moon Lee, the Chairman of AmBex Venture Partners, the students presented their team business ideas such as an AI-assisted, noise-canceling pillow devised for better sleep, a metaverse dating application, an XR virtual conferencing system, and an AI language tutoring application to the entice global investors’ curiosity. The invited investors, majorly based in Silicon Valley, commented that all the presentation was very exciting, and the level of pitches was beyond the expectation considering that the students have given only two weeks.
Ms. Seunghee Lee of the team “Bored KAIST Yacht Club”, which was awarded the first prize, explained, “our item, called ‘Meta-Everland’, is a service that offers real-time dating experiences similar to off-line dates. The GESC taught me that anybody can launch a startup as long as they are willing. Developing a business model from ideation and taking it to the actual pitching was challenging, but it was a very thrilling experience at the same time.” Lee added, “Most importantly, over the course of the program and the final pitch, I found out that an interesting idea can attract investors interest even at a very early stage of the launching.”
Mr. Byunghoon Hwang, a student who attended the program said, “Having learned the thoughts and attitudes the people at the front line of Silicon Valley, my views on career and launching of a start-up have been expanded a lot.”
Ms. Marina Mondragon, another attendee at the program, also said that the program was very meaningful because she was able to learn the difference between the ecosystem for the new start-up businesses at Korea and at Silicon Valley through her talks with the CEOs at Silicon Valley.
The program was co-organized by the Center for Global Strategies and Planning at KAIST International Office and Startup of KAIST. Dr. Man-Sung Yim, the Associate Vice President for KAIST International Office, who guided students in Silicon Valley, said, “I believe the GESC program broadened the views and entrepreneurial mindset of students. After joining this program, students stepped forward to become a founder of startups.” In addition, Dr. Young-Tae Kim, the Associate Vice President of the Institute for Startup KAIST, addressed “Startup KAIST will support business items founded via the program through various other programs in order to enhance their competitiveness in the global market.”
The GSP and Startup KAIST will continuously revamp the program by selecting distinguished fellows to join the program and coming up with innovative startup items.
Profile:
Sooa Lee, Ph.D.
Research Assistant Professor
slee900@kaist.ac.kr
Center for Global Strategies and Planning
Office of Global Initiatives
KAIST International Office
https://io.kaist.ac.kr
Korea Advanced Institute of Science and Technology (KAIST)Daejeon, Republic of Korea
2022.07.05 View 13651
The 1st Global Entrepreneurship Summer Camp bridges KAIST and Silicon Valley, US
Twenty KAIST students gave a go at selling their business ideas to investors at Silicon Valley on the “Pitch Day” at 2022 Global Entrepreneurship Summer Camp.
From Tuesday, June 21 to Monday, July 4, 2022, KAIST held the first Global Entrepreneurship Summer Camp (GESC).
The 2022 GESC, which was organized in collaboration with Stanford Technology Ventures Program (STVP), KOTRA Silicon Valley IT Center, and KAIST Alumni at Silicon Valley, was a pilot program that offered opportunities of experiencing and learning about the cases of startup companies in Silicon Valley and a chance to expand businesses to Silicon Valley through networking.
Twenty KAIST students, including pre-startup entrepreneurs and students interested in global entrepreneurship with less than one year of business experience were selected. The first week of the program was organized by Startup KAIST while the second week program was organized by the Center for Global Strategies and Planning (GSP) at KAIST in collaboration with the Stanford Technology Venture Program (STVP), KAIST Alumni at Silicon Valley, and KOTRA at Silicon Valley.
Dr. Mo-Yun Lei Fong, the Executive Director of STVP, said, “The program offered an opportunity for us to realize our vision of empowering aspiring entrepreneurs to become global citizens who create and scale responsible innovation. By collaborating with KAIST and offering entrepreneurial insights to Korean students, we are able to have a positive impact on a global scale.” Mo added, “The program also enabled STVP to build bridges, learn from the students, and refine our culturally relevant curriculum by understanding Korean culture and ideas.”
On the “Pitch Day” on July 1, following a special talk by Dr. Chong-Moon Lee, the Chairman of AmBex Venture Partners, the students presented their team business ideas such as an AI-assisted, noise-canceling pillow devised for better sleep, a metaverse dating application, an XR virtual conferencing system, and an AI language tutoring application to the entice global investors’ curiosity. The invited investors, majorly based in Silicon Valley, commented that all the presentation was very exciting, and the level of pitches was beyond the expectation considering that the students have given only two weeks.
Ms. Seunghee Lee of the team “Bored KAIST Yacht Club”, which was awarded the first prize, explained, “our item, called ‘Meta-Everland’, is a service that offers real-time dating experiences similar to off-line dates. The GESC taught me that anybody can launch a startup as long as they are willing. Developing a business model from ideation and taking it to the actual pitching was challenging, but it was a very thrilling experience at the same time.” Lee added, “Most importantly, over the course of the program and the final pitch, I found out that an interesting idea can attract investors interest even at a very early stage of the launching.”
Mr. Byunghoon Hwang, a student who attended the program said, “Having learned the thoughts and attitudes the people at the front line of Silicon Valley, my views on career and launching of a start-up have been expanded a lot.”
Ms. Marina Mondragon, another attendee at the program, also said that the program was very meaningful because she was able to learn the difference between the ecosystem for the new start-up businesses at Korea and at Silicon Valley through her talks with the CEOs at Silicon Valley.
The program was co-organized by the Center for Global Strategies and Planning at KAIST International Office and Startup of KAIST. Dr. Man-Sung Yim, the Associate Vice President for KAIST International Office, who guided students in Silicon Valley, said, “I believe the GESC program broadened the views and entrepreneurial mindset of students. After joining this program, students stepped forward to become a founder of startups.” In addition, Dr. Young-Tae Kim, the Associate Vice President of the Institute for Startup KAIST, addressed “Startup KAIST will support business items founded via the program through various other programs in order to enhance their competitiveness in the global market.”
The GSP and Startup KAIST will continuously revamp the program by selecting distinguished fellows to join the program and coming up with innovative startup items.
Profile:
Sooa Lee, Ph.D.
Research Assistant Professor
slee900@kaist.ac.kr
Center for Global Strategies and Planning
Office of Global Initiatives
KAIST International Office
https://io.kaist.ac.kr
Korea Advanced Institute of Science and Technology (KAIST)Daejeon, Republic of Korea
2022.07.05 View 13651