department+of+Chemical+and+Biomolecular+Engineering
-
 Structural Insight into the Molecular Mechanism of PET Degradation
A KAIST metabolic engineering research team has newly suggested a molecular mechanism showing superior degradability of poly ethylene terephthalate (PET).
This is the first report to simultaneously determine the 3D crystal structure of Ideonella sakaiensis PETase and develop the new variant with enhanced PET degradation.
Recently, diverse research projects are working to address the non-degradability of materials. A poly ethylene terephthalate (PET)-degrading bacterium called Ideonella sakaiensis was recently identified for the possible degradation and recycling of PET by Japanese team in Science journal (Yoshida et al., 2016). However, the detailed molecular mechanism of PET degradation has not been yet identified.
The team under Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering and the team under Professor Kyung-Jin Kim of the Department of Biotechnology at Kyungpook National University conducted this research. The findings were published in Nature Communications on January 26.
This research predicts a special molecular mechanism based on the docking simulation between PETase and a PET alternative mimic substrate. Furthermore, they succeeded in constructing the variant for IsPETase with enhanced PET-degrading activity using structural-based protein engineering.
It is expected that the new approaches taken in this research can be background for further study of other enzymes capable of degrading not only PET but other plastics as well.
PET is very important source in our daily lives. However, PET after use causes tremendous contamination issues to our environment due to its non-biodegradability, which has been a major advantage of PET. Conventionally, PET is disposed of in landfills, using incineration, and sometimes recycling using chemical methods, which induces additional environmental pollution. Therefore, a new development for highly-efficient PET degrading enzymes is essential to degrade PET using bio-based eco-friendly methods.
Recently, a new bacterial species, Ideonella sakaiensis, which can use PET as a carbon source, was isolated. The PETase of I. sakaiensis (IsPETase) can degrade PET with relatively higher success than other PET-degrading enzymes. However, the detailed enzyme mechanism has not been elucidated, hindering further studies.
The research teams investigated how the substrate binds to the enzyme and which differences in enzyme structure result in significantly higher PET degrading activity compared with other cutinases and esterases, which make IsPETase highly attractive for industrial applications toward PET waste recycling.
Based on the 3D structure and related biochemical studies, they successfully predicted the reasons for extraordinary PET degrading activity of IsPETase and suggested other enzymes that can degrade PET with a newly-classified phylogenetic tree. The team proposed that 4 MHET moieties are the most properly matched substrates due to a cleft on structure even with the 10-20-mers for PET. This is meaningful in that it is the first docking simulation between PETase and PET, not its monomer.
Furthermore, they succeeded in developing a new variant with much higher PET-degrading activity using a crystal structure of this variant to show that the changed structure is better to accommodate PET substrates than wild type PETase, which will lead to developing further superior enzymes and constructing platforms for microbial plastic recycling.
Professor Lee said, “Environmental pollution from plastics remains one of the greatest challenges worldwide with the increasing consumption of plastics. We successfully constructed a new superior PET-degrading variant with the determination of a crystal structure of PETase and its degrading molecular mechanism. This novel technology will help further studies to engineer more superior enzymes with high efficiency in degrading. This will be the subject of our team’s ongoing research projects to address the global environmental pollution problem for next generation.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) from the Ministry of Science and ICT through the National Research Foundation of Korea. Further Contact: Dr. Sang Yup Lee, Distinguished Professor, KAIST, Daejeon, Korea (leesy@kaist.ac.kr, +82-42-350-3930)
(Figure: Structural insight into the molecular mechanism of poly(ethylene terephthalate) degradation and the phylogenetic tree of possible PET degrading enzymes. This schematic diagram shows the overall conceptualization for structural insight into the molecular mechanism of poly (ethylene terephthalate) degradation and the phylogenetic tree of possible PET degrading enzymes.)
2018.01.31 View 8367
Structural Insight into the Molecular Mechanism of PET Degradation
A KAIST metabolic engineering research team has newly suggested a molecular mechanism showing superior degradability of poly ethylene terephthalate (PET).
This is the first report to simultaneously determine the 3D crystal structure of Ideonella sakaiensis PETase and develop the new variant with enhanced PET degradation.
Recently, diverse research projects are working to address the non-degradability of materials. A poly ethylene terephthalate (PET)-degrading bacterium called Ideonella sakaiensis was recently identified for the possible degradation and recycling of PET by Japanese team in Science journal (Yoshida et al., 2016). However, the detailed molecular mechanism of PET degradation has not been yet identified.
The team under Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering and the team under Professor Kyung-Jin Kim of the Department of Biotechnology at Kyungpook National University conducted this research. The findings were published in Nature Communications on January 26.
This research predicts a special molecular mechanism based on the docking simulation between PETase and a PET alternative mimic substrate. Furthermore, they succeeded in constructing the variant for IsPETase with enhanced PET-degrading activity using structural-based protein engineering.
It is expected that the new approaches taken in this research can be background for further study of other enzymes capable of degrading not only PET but other plastics as well.
PET is very important source in our daily lives. However, PET after use causes tremendous contamination issues to our environment due to its non-biodegradability, which has been a major advantage of PET. Conventionally, PET is disposed of in landfills, using incineration, and sometimes recycling using chemical methods, which induces additional environmental pollution. Therefore, a new development for highly-efficient PET degrading enzymes is essential to degrade PET using bio-based eco-friendly methods.
Recently, a new bacterial species, Ideonella sakaiensis, which can use PET as a carbon source, was isolated. The PETase of I. sakaiensis (IsPETase) can degrade PET with relatively higher success than other PET-degrading enzymes. However, the detailed enzyme mechanism has not been elucidated, hindering further studies.
The research teams investigated how the substrate binds to the enzyme and which differences in enzyme structure result in significantly higher PET degrading activity compared with other cutinases and esterases, which make IsPETase highly attractive for industrial applications toward PET waste recycling.
Based on the 3D structure and related biochemical studies, they successfully predicted the reasons for extraordinary PET degrading activity of IsPETase and suggested other enzymes that can degrade PET with a newly-classified phylogenetic tree. The team proposed that 4 MHET moieties are the most properly matched substrates due to a cleft on structure even with the 10-20-mers for PET. This is meaningful in that it is the first docking simulation between PETase and PET, not its monomer.
Furthermore, they succeeded in developing a new variant with much higher PET-degrading activity using a crystal structure of this variant to show that the changed structure is better to accommodate PET substrates than wild type PETase, which will lead to developing further superior enzymes and constructing platforms for microbial plastic recycling.
Professor Lee said, “Environmental pollution from plastics remains one of the greatest challenges worldwide with the increasing consumption of plastics. We successfully constructed a new superior PET-degrading variant with the determination of a crystal structure of PETase and its degrading molecular mechanism. This novel technology will help further studies to engineer more superior enzymes with high efficiency in degrading. This will be the subject of our team’s ongoing research projects to address the global environmental pollution problem for next generation.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) from the Ministry of Science and ICT through the National Research Foundation of Korea. Further Contact: Dr. Sang Yup Lee, Distinguished Professor, KAIST, Daejeon, Korea (leesy@kaist.ac.kr, +82-42-350-3930)
(Figure: Structural insight into the molecular mechanism of poly(ethylene terephthalate) degradation and the phylogenetic tree of possible PET degrading enzymes. This schematic diagram shows the overall conceptualization for structural insight into the molecular mechanism of poly (ethylene terephthalate) degradation and the phylogenetic tree of possible PET degrading enzymes.)
2018.01.31 View 8367 -
 One-Step Production of Aromatic Polyesters by E. coli Strains
KAIST systems metabolic engineers defined a novel strategy for microbial aromatic polyesters production fused with synthetic biology from renewable biomass. The team of Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering produced aromatic polyesters from Escherichia coli (E. coli) strains by applying microbial fermentation, employing direct microbial fermentation from renewable feedstock carbohydrates.
This is the first report to determine a platform strain of engineered E. coli capable of producing environmentally friendly aromatic polyesters. This engineered E. coli strain, if desired, has the potential to be used as a platform strain capable of producing various high-valued aromatic polyesters from renewable biomass. This research was published in Nature Communications on January 8.
Conventionally, aromatic polyesters boast solid strength and heat stability so that there has been a great deal of interest in fermentative production of aromatic polyesters from renewable non-food biomass, but without success.
However, aromatic polyesters are only made by feeding the cells with corresponding aromatic monomers as substrates, and have not been produced by direct fermentation from renewable feedstock carbohydrates such as glucose.
To address this issue, the team prescribed the detailed procedure for aromatic polyester production through identifying CoA-transferase that activates phenylalkanoates into their corresponding CoA derivatives. In this process, researchers employed metabolic engineering of E. coli to produce phenylalkanoates from glucose based on genome-scale metabolic flux analysis. In particular, the KAIST team made a modulation of gene expression to produce various aromatic polyesters having different monomer fractions.
The research team successfully produced aromatic polyesters, a non-natural polymer using the strategy that combines systems metabolic engineering and synthetic biology. They succeeded in biosynthesis of various kinds of aromatic polyesters through the system, thus proving the technical excellence of the environmentally friendly biosynthetic system of this research. Furthermore, his team also proved the potential of expanding the range of aromatic polyesters from renewable resources, which is expected to play an important role in the bio-plastic industry.
Professor Lee said, “An eco-friendly and sustainable chemical industry is the key global agenda every nation faces. We are making a research focus to a biochemical industry free from petroleum dependence, and conducting diverse research activities to address the issue. This novel technology we are presenting will serve as an opportunity to advance the biochemical industry moving forward.”
This work was supported by the Intelligent Synthetic Biology Center through the Global Frontier Project (2011-0031963) and also by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) from the Ministry of Science and ICT through the National Research Foundation of Korea.
Figure: Biosynthesis of aromatic polyesters by metabolically engineered E. coli.This schematic diagram shows the overall conceptualization of how metabolically engineered E. coli produced aromatic polyesters from glucose.
2018.01.09 View 6783
One-Step Production of Aromatic Polyesters by E. coli Strains
KAIST systems metabolic engineers defined a novel strategy for microbial aromatic polyesters production fused with synthetic biology from renewable biomass. The team of Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering produced aromatic polyesters from Escherichia coli (E. coli) strains by applying microbial fermentation, employing direct microbial fermentation from renewable feedstock carbohydrates.
This is the first report to determine a platform strain of engineered E. coli capable of producing environmentally friendly aromatic polyesters. This engineered E. coli strain, if desired, has the potential to be used as a platform strain capable of producing various high-valued aromatic polyesters from renewable biomass. This research was published in Nature Communications on January 8.
Conventionally, aromatic polyesters boast solid strength and heat stability so that there has been a great deal of interest in fermentative production of aromatic polyesters from renewable non-food biomass, but without success.
However, aromatic polyesters are only made by feeding the cells with corresponding aromatic monomers as substrates, and have not been produced by direct fermentation from renewable feedstock carbohydrates such as glucose.
To address this issue, the team prescribed the detailed procedure for aromatic polyester production through identifying CoA-transferase that activates phenylalkanoates into their corresponding CoA derivatives. In this process, researchers employed metabolic engineering of E. coli to produce phenylalkanoates from glucose based on genome-scale metabolic flux analysis. In particular, the KAIST team made a modulation of gene expression to produce various aromatic polyesters having different monomer fractions.
The research team successfully produced aromatic polyesters, a non-natural polymer using the strategy that combines systems metabolic engineering and synthetic biology. They succeeded in biosynthesis of various kinds of aromatic polyesters through the system, thus proving the technical excellence of the environmentally friendly biosynthetic system of this research. Furthermore, his team also proved the potential of expanding the range of aromatic polyesters from renewable resources, which is expected to play an important role in the bio-plastic industry.
Professor Lee said, “An eco-friendly and sustainable chemical industry is the key global agenda every nation faces. We are making a research focus to a biochemical industry free from petroleum dependence, and conducting diverse research activities to address the issue. This novel technology we are presenting will serve as an opportunity to advance the biochemical industry moving forward.”
This work was supported by the Intelligent Synthetic Biology Center through the Global Frontier Project (2011-0031963) and also by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) from the Ministry of Science and ICT through the National Research Foundation of Korea.
Figure: Biosynthesis of aromatic polyesters by metabolically engineered E. coli.This schematic diagram shows the overall conceptualization of how metabolically engineered E. coli produced aromatic polyesters from glucose.
2018.01.09 View 6783 -
 Expanding Gas Storage Capacity of Nanoporous Materials
A KAIST research team led by Professor Jihan Kim of the Department of Chemical and Biomolecular Engineering has successfully proposed a rational defect engineering methodology that can greatly enhance the gas storage capacity of nanoporous materials. The team conducted a high-throughput computational screening of a large experimental metal-organic framework database to identify 13 candidate materials that could experience significant methane uptake enhancement with only a small proportion of linker vacancy defects.
This research was published online on November 16 in Nature Communications, with M.S. candidate Sanggyu Chong from KAIST as the first author and post-doctorate researcher Günther Thiele from the Department of Chemistry at UC Berkeley as a contributing author.
Metal-organic frameworks, hereinafter MOF, are crystalline nanoporous materials that are comprised of metal clusters and organic linkers continuously bound together by coordination bonds. Due to their ultrahigh surface areas and pore volumes, they have been widely studied for various energy and environment applications.
Similar to other crystalline materials, MOFs are never perfectly crystalline and are likely to contain several different types of defects within their crystalline structures. Among these defects, linker vacancy defects, or the random absence of linker vacancies in their designated bonding positions, are known to be controllable by practicing careful control over the synthesis conditions.
The research team combined the concepts of rational defect engineering over the linker vacancy defects and the potential presence of inaccessible pores within MOFs to propose a methodology where controlled the introduction of linker vacancy defects could lead to a dramatic enhancement in gas adsorption and storage capacities.
The study utilized a Graphic Processing Unit (GPU) code developed by Professor Kim in a high-throughput computational screening of 12,000 experimentally synthesized MOFs to identify the structures with significant amounts of pores that were inaccessible for methane. In determining the presence of inaccessible pores, a flood-fill algorithm was performed over the energy-low regions of the structure, which is the same algorithm used for filling an area with color in Microsoft Paint.
For the MOFs with significant amounts of inaccessible pores, as determined from the screening, the research team emulated linker vacancy defects in their crystalline structures so that the previously inaccessible pores would be newly merged into the main adsorption channel with the introduction of defects for additional surface area and pore volume available for adsorption. The research team successfully identified 13 structures that would experience up to a 55.56% increase in their methane uptake with less than 8.33% of the linker vacancy defects.
The research team believes that this rational defect engineering scheme can be further utilized for many other applications in areas such as selective adsorption of an adsorbate from a gas mixture and the semi-permanent capture of gas molecules.
This research was conducted with the support of the Mid-career Research Program of the National Research Foundation of Korea.
Figure1. A diagram for flood fill algorithm and example of identification of inaccessible regions within the MOFs, using the flood fill algorithm
Figure2. Methane energy contours before and after detect introduction
2017.12.04 View 7147
Expanding Gas Storage Capacity of Nanoporous Materials
A KAIST research team led by Professor Jihan Kim of the Department of Chemical and Biomolecular Engineering has successfully proposed a rational defect engineering methodology that can greatly enhance the gas storage capacity of nanoporous materials. The team conducted a high-throughput computational screening of a large experimental metal-organic framework database to identify 13 candidate materials that could experience significant methane uptake enhancement with only a small proportion of linker vacancy defects.
This research was published online on November 16 in Nature Communications, with M.S. candidate Sanggyu Chong from KAIST as the first author and post-doctorate researcher Günther Thiele from the Department of Chemistry at UC Berkeley as a contributing author.
Metal-organic frameworks, hereinafter MOF, are crystalline nanoporous materials that are comprised of metal clusters and organic linkers continuously bound together by coordination bonds. Due to their ultrahigh surface areas and pore volumes, they have been widely studied for various energy and environment applications.
Similar to other crystalline materials, MOFs are never perfectly crystalline and are likely to contain several different types of defects within their crystalline structures. Among these defects, linker vacancy defects, or the random absence of linker vacancies in their designated bonding positions, are known to be controllable by practicing careful control over the synthesis conditions.
The research team combined the concepts of rational defect engineering over the linker vacancy defects and the potential presence of inaccessible pores within MOFs to propose a methodology where controlled the introduction of linker vacancy defects could lead to a dramatic enhancement in gas adsorption and storage capacities.
The study utilized a Graphic Processing Unit (GPU) code developed by Professor Kim in a high-throughput computational screening of 12,000 experimentally synthesized MOFs to identify the structures with significant amounts of pores that were inaccessible for methane. In determining the presence of inaccessible pores, a flood-fill algorithm was performed over the energy-low regions of the structure, which is the same algorithm used for filling an area with color in Microsoft Paint.
For the MOFs with significant amounts of inaccessible pores, as determined from the screening, the research team emulated linker vacancy defects in their crystalline structures so that the previously inaccessible pores would be newly merged into the main adsorption channel with the introduction of defects for additional surface area and pore volume available for adsorption. The research team successfully identified 13 structures that would experience up to a 55.56% increase in their methane uptake with less than 8.33% of the linker vacancy defects.
The research team believes that this rational defect engineering scheme can be further utilized for many other applications in areas such as selective adsorption of an adsorbate from a gas mixture and the semi-permanent capture of gas molecules.
This research was conducted with the support of the Mid-career Research Program of the National Research Foundation of Korea.
Figure1. A diagram for flood fill algorithm and example of identification of inaccessible regions within the MOFs, using the flood fill algorithm
Figure2. Methane energy contours before and after detect introduction
2017.12.04 View 7147 -
 Technology Detecting RNase Activity
(Ph.D. candidate Chang Yeol Lee)
A KAIST research team of Professor Hyun Gyu Park at Department of Chemical and Biomolecular Engineering developed a new technology to detect the activity of RNase H, a RNA degrading enzyme. The team used highly efficient signal amplification reaction termed catalytic hairpin assembly (CHA) to effectively analyze the RNase H activity. Considering that RNase H is required in the proliferation of retroviruses such as HIV, this research finding could contribute to AIDS treatments in the future, researchers say.
This study led by Ph.D. candidates Chang Yeol Lee and Hyowon Jang was chosen as the cover for Nanoscale (Issue 42, 2017) published in 14 November.
The existing techniques to detect RNase H require expensive fluorophore and quencher, and involve complex implementation. Further, there is no way to amplify the signal, leading to low detection efficiency overall. The team utilized CHA technology to overcome these limitations. CHA amplifies detection signal to allow more sensitive RNase H activity assay.
The team designed the reaction system so that the product of CHA reaction has G-quadruplex structures, which is suitable to generate fluorescence. By using fluorescent molecules that bind to G-quadruplexes to generate strong fluorescence, the team could develop high performance RNase H detection method that overcomes the limitations of existing techniques. Further, this technology could screen inhibitors of RNase H activity.
The team expects that the research finding could contribute to AIDS treatment. AIDS is disease caused by HIV, a retrovirus that utilizes reverse transcription, during which RNA is converted to DNA. RNase H is essential for reverse transcription in HIV, and thus inhibition of RNase H could in turn inhibit transcription of HIV DNA.
Professor Park said, “This technology is applicable to detect various enzyme activities, as well as RNase H activity.” He continued, “I hope this technology could be widely used in research on enzyme related diseases.”
This study was funded by Global Frontier project and Mid-career Researcher Support project of the Ministry of Science and ICT.
2017.11.28 View 6197
Technology Detecting RNase Activity
(Ph.D. candidate Chang Yeol Lee)
A KAIST research team of Professor Hyun Gyu Park at Department of Chemical and Biomolecular Engineering developed a new technology to detect the activity of RNase H, a RNA degrading enzyme. The team used highly efficient signal amplification reaction termed catalytic hairpin assembly (CHA) to effectively analyze the RNase H activity. Considering that RNase H is required in the proliferation of retroviruses such as HIV, this research finding could contribute to AIDS treatments in the future, researchers say.
This study led by Ph.D. candidates Chang Yeol Lee and Hyowon Jang was chosen as the cover for Nanoscale (Issue 42, 2017) published in 14 November.
The existing techniques to detect RNase H require expensive fluorophore and quencher, and involve complex implementation. Further, there is no way to amplify the signal, leading to low detection efficiency overall. The team utilized CHA technology to overcome these limitations. CHA amplifies detection signal to allow more sensitive RNase H activity assay.
The team designed the reaction system so that the product of CHA reaction has G-quadruplex structures, which is suitable to generate fluorescence. By using fluorescent molecules that bind to G-quadruplexes to generate strong fluorescence, the team could develop high performance RNase H detection method that overcomes the limitations of existing techniques. Further, this technology could screen inhibitors of RNase H activity.
The team expects that the research finding could contribute to AIDS treatment. AIDS is disease caused by HIV, a retrovirus that utilizes reverse transcription, during which RNA is converted to DNA. RNase H is essential for reverse transcription in HIV, and thus inhibition of RNase H could in turn inhibit transcription of HIV DNA.
Professor Park said, “This technology is applicable to detect various enzyme activities, as well as RNase H activity.” He continued, “I hope this technology could be widely used in research on enzyme related diseases.”
This study was funded by Global Frontier project and Mid-career Researcher Support project of the Ministry of Science and ICT.
2017.11.28 View 6197 -
 Highly Flexible Organic Flash Memory for Foldable and Disposable Electronics
A KAIST team reported ultra-flexible organic flash memory that is bendable down to a radius of 300μm. The memory exhibits a significantly-long projected retention rate with a programming voltage on par with the present industrial standards.
A joint research team led by Professor Seunghyup Yoo of the School of Electrical Engineering and Professor Sung Gap Im of the Department of Chemical and Biomolecular Engineering said that their memory technology can be applied to non-conventional substrates, such as plastics and papers, to demonstrate its feasibility over a wide range of applications.
With Dr. Seungwon Lee and Dr. Hanul Moon playing the role of leading authors, the research was published in Nature Communications on September 28.
Flash memory is a non-volatile, transistor-based data-storage device that has become essential in most electronic systems in daily life. With straightforward operation mechanisms and easy integration into NAND or NOR array architecture, flash memory has been established as the most successful and dominant non-volatile memory technology by far.
Despite promising demonstrations in the early stages of organic electronics, the overall progress in this field has been far slower than that of thin-film transistors (TFTs) or other devices based on flexible materials. It has been challenging, in particular, to develop flash memory that simultaneously exhibits a significant level of flexibility and performance. This is mainly due to the scarcity of flexible dielectric layers, which are responsible for the tunneling and blocking of charges.
The solution processing used for the preparation of most of the polymeric dielectric layers also makes it difficult to use them in flash memory due to the complexity involved in the formation of the bilayer dielectric structure, which is the key to flash memory operations.
The research team tried to overcome these hurdles and realize highly flexible flash memory by employing thin polymeric insulators grown with initiated chemical vapor deposition (iCVD), a vapor-phase growth technique for polymers that was previously shown to be promising for the fabrication of flexible TFTs. It was further shown that these iCVD-based polymeric insulators, when coupled with rational device design and material choice, can make a significant contribution to flash memory as well.
Memory using conventional polymer insulating films has often required a voltage as high as 100 V (volt) in order to attain long memory retention. If the device is made to operate at a low voltage, the short retention period of less than a month was problematic.
The KAIST team produced flash memory with programming voltages around 10 V and a projected data retention time of over 10 years, while maintaining its memory performance even at a mechanical strain of 2.8%. This is a significant improvement over the existing inorganic insulation layer-based flash memory that allowed only a 1% strain.
The team demonstrated the virtually foldable memory devices by fabricating the proposed flash memory on a 6-micrometer-thick ultrathin plastic film. In addition, it succeeded in producing them on printing paper, opening a way for disposable smart electronic products such as electronic paper and electronic business card.
Professor Yoo said, " This study well illustrates that even highly flexible flash memory can be made to have a practically viable level of performance, so that it contributes to full-fledged wearable electronic devices and smart electronic paper."
(Figure 1. Structure of flexible flash memory )
(Figure 2. Foldable flash memory)
2017.11.06 View 8562
Highly Flexible Organic Flash Memory for Foldable and Disposable Electronics
A KAIST team reported ultra-flexible organic flash memory that is bendable down to a radius of 300μm. The memory exhibits a significantly-long projected retention rate with a programming voltage on par with the present industrial standards.
A joint research team led by Professor Seunghyup Yoo of the School of Electrical Engineering and Professor Sung Gap Im of the Department of Chemical and Biomolecular Engineering said that their memory technology can be applied to non-conventional substrates, such as plastics and papers, to demonstrate its feasibility over a wide range of applications.
With Dr. Seungwon Lee and Dr. Hanul Moon playing the role of leading authors, the research was published in Nature Communications on September 28.
Flash memory is a non-volatile, transistor-based data-storage device that has become essential in most electronic systems in daily life. With straightforward operation mechanisms and easy integration into NAND or NOR array architecture, flash memory has been established as the most successful and dominant non-volatile memory technology by far.
Despite promising demonstrations in the early stages of organic electronics, the overall progress in this field has been far slower than that of thin-film transistors (TFTs) or other devices based on flexible materials. It has been challenging, in particular, to develop flash memory that simultaneously exhibits a significant level of flexibility and performance. This is mainly due to the scarcity of flexible dielectric layers, which are responsible for the tunneling and blocking of charges.
The solution processing used for the preparation of most of the polymeric dielectric layers also makes it difficult to use them in flash memory due to the complexity involved in the formation of the bilayer dielectric structure, which is the key to flash memory operations.
The research team tried to overcome these hurdles and realize highly flexible flash memory by employing thin polymeric insulators grown with initiated chemical vapor deposition (iCVD), a vapor-phase growth technique for polymers that was previously shown to be promising for the fabrication of flexible TFTs. It was further shown that these iCVD-based polymeric insulators, when coupled with rational device design and material choice, can make a significant contribution to flash memory as well.
Memory using conventional polymer insulating films has often required a voltage as high as 100 V (volt) in order to attain long memory retention. If the device is made to operate at a low voltage, the short retention period of less than a month was problematic.
The KAIST team produced flash memory with programming voltages around 10 V and a projected data retention time of over 10 years, while maintaining its memory performance even at a mechanical strain of 2.8%. This is a significant improvement over the existing inorganic insulation layer-based flash memory that allowed only a 1% strain.
The team demonstrated the virtually foldable memory devices by fabricating the proposed flash memory on a 6-micrometer-thick ultrathin plastic film. In addition, it succeeded in producing them on printing paper, opening a way for disposable smart electronic products such as electronic paper and electronic business card.
Professor Yoo said, " This study well illustrates that even highly flexible flash memory can be made to have a practically viable level of performance, so that it contributes to full-fledged wearable electronic devices and smart electronic paper."
(Figure 1. Structure of flexible flash memory )
(Figure 2. Foldable flash memory)
2017.11.06 View 8562 -
 Platinum Single Atom Catalysts for 'Direct Formic Acid Fuel Cells'
(Professor Hyunjoo Lee (left) and Ph.D. candidate Jiwhan Kim)
A research team co-led by Professor Hyunjoo Lee at the Department of Chemical and Biomolecular Engineering at KAIST and Professor Jeong Woo Han from the University of Seoul synthesized highly stable high-Pt-content single atom catalysts for direct formic acid fuel cells. The amount of platinum can be reduced to 1/10 of that of conventional platinum nanoparticle catalysts.
Platinum (Pt) catalysts have been used in various catalytic reactions due to their high activity and stability. However, because Pt is rare and expensive, it is important to reduce the amount of Pt used. Pt single atom catalysts can reduce the size of the Pt particles to the size of an atom. Thus, the cost of Pt catalysts can be minimized because all of the Pt atoms can participate in the catalytic reactions. Additionally, single atom catalysts have no ensemble site in which two or more atoms are attached, and thus, the reaction selectivity is different from that of nanoparticle catalysts.
Despite these advantages, single atom catalysts are easily aggregated and less stable due to their low coordination number and high surface free energy. It is difficult to develop a single atom catalyst with high content and high stability, and thus, its application in practical devices is limited.
Direct formic acid fuel cells can be an energy source for next-generation portable devices because liquid formic acid as a fuel is safer and easier to store and transport than high-pressure hydrogen gas.
To improve the stability of Pt single atom catalysts, Professor Lee’s group developed a Pt-Sn single atom alloy structure on an antimony-doped tin oxide (ATO) support. This structure has been proven by computational calculations which show that Pt single atoms substitute antimony sites in the antimony-tin alloy structure and are thermodynamically stable. This catalyst has been shown to have a higher activity up to 50 times per weight of Pt than that of the commercial catalyst, Pt/C, in the oxidation of formic acid, and the stability of the catalyst was also remarkably high.
Professor Lee’s group also used a single atomic catalyst in a 'direct formic acid fuel cell’ consisting of membranes and electrodes. It is the first attempt to apply a single atomic catalyst to a full cell. In this case, an output similar to that of the commercial catalyst could be obtained by using 1/10 of the platinum compared to the commercial Pt/C catalyst.
Ph.D. candidate Jiwhan Kim from KAIST was the first author of the research. This research was published online on September 11 in Advanced Energy Materials.
This research was carried out with the support of the Samsung Electronics Future Technology Development Center.
(Figure 1. Concept photograph for Pt single atom catalysts.)
(Figure 2. Pt single atom catalysts by HAADF-STEM analysis (bright white circles))
2017.10.31 View 6937
Platinum Single Atom Catalysts for 'Direct Formic Acid Fuel Cells'
(Professor Hyunjoo Lee (left) and Ph.D. candidate Jiwhan Kim)
A research team co-led by Professor Hyunjoo Lee at the Department of Chemical and Biomolecular Engineering at KAIST and Professor Jeong Woo Han from the University of Seoul synthesized highly stable high-Pt-content single atom catalysts for direct formic acid fuel cells. The amount of platinum can be reduced to 1/10 of that of conventional platinum nanoparticle catalysts.
Platinum (Pt) catalysts have been used in various catalytic reactions due to their high activity and stability. However, because Pt is rare and expensive, it is important to reduce the amount of Pt used. Pt single atom catalysts can reduce the size of the Pt particles to the size of an atom. Thus, the cost of Pt catalysts can be minimized because all of the Pt atoms can participate in the catalytic reactions. Additionally, single atom catalysts have no ensemble site in which two or more atoms are attached, and thus, the reaction selectivity is different from that of nanoparticle catalysts.
Despite these advantages, single atom catalysts are easily aggregated and less stable due to their low coordination number and high surface free energy. It is difficult to develop a single atom catalyst with high content and high stability, and thus, its application in practical devices is limited.
Direct formic acid fuel cells can be an energy source for next-generation portable devices because liquid formic acid as a fuel is safer and easier to store and transport than high-pressure hydrogen gas.
To improve the stability of Pt single atom catalysts, Professor Lee’s group developed a Pt-Sn single atom alloy structure on an antimony-doped tin oxide (ATO) support. This structure has been proven by computational calculations which show that Pt single atoms substitute antimony sites in the antimony-tin alloy structure and are thermodynamically stable. This catalyst has been shown to have a higher activity up to 50 times per weight of Pt than that of the commercial catalyst, Pt/C, in the oxidation of formic acid, and the stability of the catalyst was also remarkably high.
Professor Lee’s group also used a single atomic catalyst in a 'direct formic acid fuel cell’ consisting of membranes and electrodes. It is the first attempt to apply a single atomic catalyst to a full cell. In this case, an output similar to that of the commercial catalyst could be obtained by using 1/10 of the platinum compared to the commercial Pt/C catalyst.
Ph.D. candidate Jiwhan Kim from KAIST was the first author of the research. This research was published online on September 11 in Advanced Energy Materials.
This research was carried out with the support of the Samsung Electronics Future Technology Development Center.
(Figure 1. Concept photograph for Pt single atom catalysts.)
(Figure 2. Pt single atom catalysts by HAADF-STEM analysis (bright white circles))
2017.10.31 View 6937 -
 Development of a Highly-Accurate Computational Model of Human Metabolism
A research team from KAIST developed a computational framework that enables the reconstruction of a comprehensive computational model of human metabolism, which allows for an accurate prediction of personal metabolic features (or phenotypes).
Understanding personal metabolic phenotypes allows us to design effective therapeutic strategies for various chronic and infectious diseases. A human computational model called the genome-scale metabolic model (GEM) contains information on thousands of metabolic genes and their corresponding reactions and metabolites, and has played an important role in predicting metabolic phenotypes. Although several versions of human GEMs have been released, they had room for further development, especially as to incorporating biological information coming from a human genetics mechanism called “alternative splicing.” Alternative splicing is a genetic mechanism that allows a gene to give rise to multiple reactions, and is strongly associated with pathology.
To tackle this problem, Jae Yong Ryu (a Ph.D. student), Dr. Hyun Uk Kim (Research Fellow), and Distinguished Professor Sang Yup Lee, all from the Department of Chemical and Biomolecular Engineering at KAIST, developed a computational framework that systematically generates metabolic reactions, and adds them to the human GEM. The resulting human GEM was demonstrated to accurately predict metabolic phenotypes under varied environmental conditions. The research results were published online in Proceedings of the National Academy of Sciences (PNAS) on October 24, 2017, under the title “Framework and resource for more than 11,000 gene-transcript-protein-reaction associations in human metabolism.”
The research team first updated the biological contents of a previous version of the human GEM. The updated biological contents include metabolic genes and their corresponding metabolites and reactions. In particular, metabolic reactions catalyzed by already-known protein isoforms were additionally incorporated into the human GEM; protein isoforms are multiple variants of proteins generated from individual genes through the alternative splicing process. Each protein isoform is often responsible for the operation of a metabolic reaction. Although multiple protein isoforms generated from one gene can play different functions by having different sets of protein domains and/or subcellular localizations, such information was not properly considered in previous versions of human GEMs.
Upon the initial update of the human GEM, named Recon 2M.1, the research team subsequently implemented a computational framework that systematically generates information on Gene-Transcript-Protein-Reaction Associations (GeTPRA) in order to identify protein isoforms that were previously not identified. This framework was developed in this study. As a result of the implementation of the framework for GeTPRA, more than 11,000 GeTPRA were automatically predicted, and thoroughly validated. Additional metabolic reactions were then added to Recon 2M.1 based on the predicted GeTPRA for the previously uncharacterized protein isoforms; Recon 2M.1 was renamed Recon 2M.2 from this upgrade.
Finally, Recon 2M.2 was integrated with 446 sets of personal biological data (RNA-Seq data) in order to build patient-specific cancer models. These patient-specific cancer models were used to predict cancer metabolism activities and anticancer targets.
The development of a new version of human GEMs along with the computational framework for GeTPRA is expected to boost studies in fundamental human genetics and medicine. Model files of the human GEMs Recon 2M.1 and 2M.2, a full list of the GeTPRA and the source code for the computational framework to predict the GeTPRA are all available as part of the publication of this study.
Distinguished Professor Lee said, “The predicted GeTPRA from the computational framework is expected to serve as a guideline for future experiments on human genetics and biochemistry, whereas the resulting Recon 2M.2 can be used to predict drug targets for various human diseases.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) from the Ministry of Science and ICT through the National Research Foundation (NRF) of Korea.
(Figure 1:A scheme of Recon 2M.1 development and its use in reconstructing personal genome-scale metabolic models (GEMs). (A) A concept of alternative splicing of human genes and its use in Gene-Transcript-Protein-Reaction Associations (GeTPRA) of Recon 2M.1. (B) A procedure of systematic refinement of the Recon 2Q. Recon 2Q is one of the previously released human GEMs. Biochemically inconsistent reactions include unbalanced, artificial, blocked, and/or redundant reactions. Iterative manual curation was conducted while validating the Recon 2M.1. (C) Reconstruction of cancer patient-specific GEMs using Recon 2M.1 for further simulation studies. In this study, personal biological data (RNA-Seq data) were obtained from The Cancer Genome Atlas (TCGA; https://cancergenome.nih.gov/ ) across the ten cancer types.
(Figure 2: Computational framework for the systematic generation of Gene-Transcript-Protein-Reaction Associations (GeTPRA; red box in the flowchart). Peptide sequences of metabolic genes defined in Recon 2M.1 were retrieved from a database called Ensembl. EC numbers and subcellular localizations of all the protein isoforms of metabolic genes in Recon 2M.1 were predicted using software programs EFICAz2.5 and Wolf PSort, respectively. Information on the newly predicted GeTPRA was systematically incorporated into the Recon 2M.1, thereby resulting in Recon 2M.2.)
2017.10.25 View 8674
Development of a Highly-Accurate Computational Model of Human Metabolism
A research team from KAIST developed a computational framework that enables the reconstruction of a comprehensive computational model of human metabolism, which allows for an accurate prediction of personal metabolic features (or phenotypes).
Understanding personal metabolic phenotypes allows us to design effective therapeutic strategies for various chronic and infectious diseases. A human computational model called the genome-scale metabolic model (GEM) contains information on thousands of metabolic genes and their corresponding reactions and metabolites, and has played an important role in predicting metabolic phenotypes. Although several versions of human GEMs have been released, they had room for further development, especially as to incorporating biological information coming from a human genetics mechanism called “alternative splicing.” Alternative splicing is a genetic mechanism that allows a gene to give rise to multiple reactions, and is strongly associated with pathology.
To tackle this problem, Jae Yong Ryu (a Ph.D. student), Dr. Hyun Uk Kim (Research Fellow), and Distinguished Professor Sang Yup Lee, all from the Department of Chemical and Biomolecular Engineering at KAIST, developed a computational framework that systematically generates metabolic reactions, and adds them to the human GEM. The resulting human GEM was demonstrated to accurately predict metabolic phenotypes under varied environmental conditions. The research results were published online in Proceedings of the National Academy of Sciences (PNAS) on October 24, 2017, under the title “Framework and resource for more than 11,000 gene-transcript-protein-reaction associations in human metabolism.”
The research team first updated the biological contents of a previous version of the human GEM. The updated biological contents include metabolic genes and their corresponding metabolites and reactions. In particular, metabolic reactions catalyzed by already-known protein isoforms were additionally incorporated into the human GEM; protein isoforms are multiple variants of proteins generated from individual genes through the alternative splicing process. Each protein isoform is often responsible for the operation of a metabolic reaction. Although multiple protein isoforms generated from one gene can play different functions by having different sets of protein domains and/or subcellular localizations, such information was not properly considered in previous versions of human GEMs.
Upon the initial update of the human GEM, named Recon 2M.1, the research team subsequently implemented a computational framework that systematically generates information on Gene-Transcript-Protein-Reaction Associations (GeTPRA) in order to identify protein isoforms that were previously not identified. This framework was developed in this study. As a result of the implementation of the framework for GeTPRA, more than 11,000 GeTPRA were automatically predicted, and thoroughly validated. Additional metabolic reactions were then added to Recon 2M.1 based on the predicted GeTPRA for the previously uncharacterized protein isoforms; Recon 2M.1 was renamed Recon 2M.2 from this upgrade.
Finally, Recon 2M.2 was integrated with 446 sets of personal biological data (RNA-Seq data) in order to build patient-specific cancer models. These patient-specific cancer models were used to predict cancer metabolism activities and anticancer targets.
The development of a new version of human GEMs along with the computational framework for GeTPRA is expected to boost studies in fundamental human genetics and medicine. Model files of the human GEMs Recon 2M.1 and 2M.2, a full list of the GeTPRA and the source code for the computational framework to predict the GeTPRA are all available as part of the publication of this study.
Distinguished Professor Lee said, “The predicted GeTPRA from the computational framework is expected to serve as a guideline for future experiments on human genetics and biochemistry, whereas the resulting Recon 2M.2 can be used to predict drug targets for various human diseases.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) from the Ministry of Science and ICT through the National Research Foundation (NRF) of Korea.
(Figure 1:A scheme of Recon 2M.1 development and its use in reconstructing personal genome-scale metabolic models (GEMs). (A) A concept of alternative splicing of human genes and its use in Gene-Transcript-Protein-Reaction Associations (GeTPRA) of Recon 2M.1. (B) A procedure of systematic refinement of the Recon 2Q. Recon 2Q is one of the previously released human GEMs. Biochemically inconsistent reactions include unbalanced, artificial, blocked, and/or redundant reactions. Iterative manual curation was conducted while validating the Recon 2M.1. (C) Reconstruction of cancer patient-specific GEMs using Recon 2M.1 for further simulation studies. In this study, personal biological data (RNA-Seq data) were obtained from The Cancer Genome Atlas (TCGA; https://cancergenome.nih.gov/ ) across the ten cancer types.
(Figure 2: Computational framework for the systematic generation of Gene-Transcript-Protein-Reaction Associations (GeTPRA; red box in the flowchart). Peptide sequences of metabolic genes defined in Recon 2M.1 were retrieved from a database called Ensembl. EC numbers and subcellular localizations of all the protein isoforms of metabolic genes in Recon 2M.1 were predicted using software programs EFICAz2.5 and Wolf PSort, respectively. Information on the newly predicted GeTPRA was systematically incorporated into the Recon 2M.1, thereby resulting in Recon 2M.2.)
2017.10.25 View 8674 -
 A Novel and Practical Fab-route for Superomniphobic Liquid-free Surfaces
(clockwise from left: Jaeho Choi, Hee Tak Kim, Shin-Hyun Kim)
A joint research team led by Professor Hee Tak Kim and Shin-Hyun Kim in the Department of Chemical and Biomolecular Engineering at KAIST developed a fabrication technology that can inexpensively produce surfaces capable of repelling liquids, including water and oil.
The team used the photofluidization of azobenzene molecule-containing polymers to generate a superomniphobic surface which can be applied for developing stain-free fabrics, non-biofouling medical tubing, and corrosion-free surfaces.
Mushroom-shaped surface textures, also called doubly re-entrant structures, are known to be the most effective surface structure that enhances resistance against liquid invasion, thereby exhibiting superior superomniphobic property.
However, the existing procedures for their fabrication are highly delicate, time-consuming, and costly. Moreover, the materials required for the fabrication are restricted to an inflexible and expensive silicon wafer, which limits the practical use of the surface.
To overcome such limitations, the research team used a different approach to fabricate the re-entrant structures called localized photofludization by using the peculiar optical phenomenon of azobenzene molecule-containing polymers (referred to as azopolymers). It is a phenomenon where an azopolymer becomes fluidized under irradiation, and the fluidization takes place locally within the thin surface layer of the azopolymer.
With this novel approach, the team facilitated the localized photofluidization in the top surface layer of azopolymer cylindrical posts, successfully reconfiguring the cylindrical posts to doubly re-entrant geometry while the fluidized thin top surface of an azopolymer is flowing down.
The structure developed by the team exhibits a superior superomniphobic property even for liquids infiltrating the surface immediately.
Moreover, the superomniphobic property can be maintained on a curved target surface because its surficial materials are based on high molecules.
Furthermore, the fabrication procedure of the structure is highly reproducible and scalable, providing a practical route to creating robust omniphobic surfaces.
Professor Hee Tak Kim said, “Not only does the novel photo-fluidization technology in this study produce superior superomniphobic surfaces, but it also possesses many practical advantages in terms of fab-procedures and material flexibility; therefore, it could greatly contribute to real uses in diverse applications.”
Professor Shin-Hyun Kim added, “The designed doubly re-entrant geometry in this study was inspired by the skin structure of springtails, insects dwelling in soil that breathe through their skin. As I carried out this research, I once again realized that humans can learn from nature to create new engineering designs.”
The paper (Jaeho Choi as a first author) was published in ACS Nano, an international journal for Nano-technology, in August.
(Schematic diagram of mushroom-shaped structure fabrication)
(SEM image of mushroom-shaped structure)
(Image of superomniphobic property of different types of liquid)
2017.09.08 View 7055
A Novel and Practical Fab-route for Superomniphobic Liquid-free Surfaces
(clockwise from left: Jaeho Choi, Hee Tak Kim, Shin-Hyun Kim)
A joint research team led by Professor Hee Tak Kim and Shin-Hyun Kim in the Department of Chemical and Biomolecular Engineering at KAIST developed a fabrication technology that can inexpensively produce surfaces capable of repelling liquids, including water and oil.
The team used the photofluidization of azobenzene molecule-containing polymers to generate a superomniphobic surface which can be applied for developing stain-free fabrics, non-biofouling medical tubing, and corrosion-free surfaces.
Mushroom-shaped surface textures, also called doubly re-entrant structures, are known to be the most effective surface structure that enhances resistance against liquid invasion, thereby exhibiting superior superomniphobic property.
However, the existing procedures for their fabrication are highly delicate, time-consuming, and costly. Moreover, the materials required for the fabrication are restricted to an inflexible and expensive silicon wafer, which limits the practical use of the surface.
To overcome such limitations, the research team used a different approach to fabricate the re-entrant structures called localized photofludization by using the peculiar optical phenomenon of azobenzene molecule-containing polymers (referred to as azopolymers). It is a phenomenon where an azopolymer becomes fluidized under irradiation, and the fluidization takes place locally within the thin surface layer of the azopolymer.
With this novel approach, the team facilitated the localized photofluidization in the top surface layer of azopolymer cylindrical posts, successfully reconfiguring the cylindrical posts to doubly re-entrant geometry while the fluidized thin top surface of an azopolymer is flowing down.
The structure developed by the team exhibits a superior superomniphobic property even for liquids infiltrating the surface immediately.
Moreover, the superomniphobic property can be maintained on a curved target surface because its surficial materials are based on high molecules.
Furthermore, the fabrication procedure of the structure is highly reproducible and scalable, providing a practical route to creating robust omniphobic surfaces.
Professor Hee Tak Kim said, “Not only does the novel photo-fluidization technology in this study produce superior superomniphobic surfaces, but it also possesses many practical advantages in terms of fab-procedures and material flexibility; therefore, it could greatly contribute to real uses in diverse applications.”
Professor Shin-Hyun Kim added, “The designed doubly re-entrant geometry in this study was inspired by the skin structure of springtails, insects dwelling in soil that breathe through their skin. As I carried out this research, I once again realized that humans can learn from nature to create new engineering designs.”
The paper (Jaeho Choi as a first author) was published in ACS Nano, an international journal for Nano-technology, in August.
(Schematic diagram of mushroom-shaped structure fabrication)
(SEM image of mushroom-shaped structure)
(Image of superomniphobic property of different types of liquid)
2017.09.08 View 7055 -
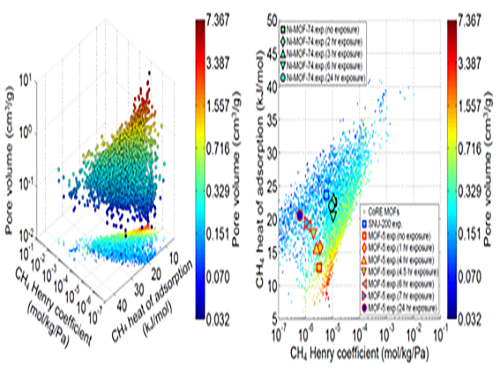 Analysis of Gas Adsorption Properties for Amorphous Porous Materials
Professor Jihan Kim from the Department of Chemical and Biomolecular Engineering at KAIST has developed a method to predict gas adsorption properties of amorphous porous materials.
Metal-organic frameworks (MOFs) have large surface area and high density of pores, making them appropriate for various energy and environmental-related applications. And although most MOFs are crystalline, these structures can deform during synthesis and/or industrial processes, leading to loss in long-range order. Unfortunately, without the structural information, existing computer simulation techniques cannot be used to model these materials.
In this research, Professor Kim’s research team demonstrated that one can replace the material properties of structurally deformed MOFs with those of crystalline MOFs to indirectly analyze/model the material properties of amorphous materials. First, the team conducted simulations on methane gas adsorption properties for over 12,000 crystalline MOFs to obtain a large training set data, and created a resulting structure-property map. Upon mapping the experimental data of amorphous MOFs onto the structure-property map, results showed that regardless of crystallinity, the gas adsorption properties of MOFs showed congruence and consistency amongst one another.
Based on these findings, selected crystalline MOFs with the most similar gas adsorption properties as the collapsed structure from the 12,000 candidates. Then, the team verified that the adsorption properties of these similar MOFs can be successfully transferred to the deformed MOFs across different temperatures and even to different gas molecules (e.g. hydrogen), demonstrating transferability of properties.
These findings allow material property prediction in porous materials such as MOFs without structural information, and the techniques here can be used to better predict and understand optimal materials for various applications including, carbon dioxide capture, gas storage and separations.
This research was conducted in collaboration with Professor Dae-Woon Lim at Kyoto University, Professor Myunghyun Paik at Seoul National University, Professor Minyoung Yoon at Gachon University, and Aadesh Harale at Saudi Arabian Oil Company. The research was published in the Proceedings of the National Academy of Sciences (PNAS) online on 10 July and the co-first authors were Ph. D. candidate WooSeok Jeong and Professor Dae-Woon Lim.
This research was funded by the Saudi Aramco-KAIST CO2 Management Center.
(Figure 1. Trends in structure - material property map and in collapsed structures)
(Figure 2. Transferability between the experimental results of collapsed MOFs and the simulation results of crystalline MOFs)
2017.07.26 View 8544
Analysis of Gas Adsorption Properties for Amorphous Porous Materials
Professor Jihan Kim from the Department of Chemical and Biomolecular Engineering at KAIST has developed a method to predict gas adsorption properties of amorphous porous materials.
Metal-organic frameworks (MOFs) have large surface area and high density of pores, making them appropriate for various energy and environmental-related applications. And although most MOFs are crystalline, these structures can deform during synthesis and/or industrial processes, leading to loss in long-range order. Unfortunately, without the structural information, existing computer simulation techniques cannot be used to model these materials.
In this research, Professor Kim’s research team demonstrated that one can replace the material properties of structurally deformed MOFs with those of crystalline MOFs to indirectly analyze/model the material properties of amorphous materials. First, the team conducted simulations on methane gas adsorption properties for over 12,000 crystalline MOFs to obtain a large training set data, and created a resulting structure-property map. Upon mapping the experimental data of amorphous MOFs onto the structure-property map, results showed that regardless of crystallinity, the gas adsorption properties of MOFs showed congruence and consistency amongst one another.
Based on these findings, selected crystalline MOFs with the most similar gas adsorption properties as the collapsed structure from the 12,000 candidates. Then, the team verified that the adsorption properties of these similar MOFs can be successfully transferred to the deformed MOFs across different temperatures and even to different gas molecules (e.g. hydrogen), demonstrating transferability of properties.
These findings allow material property prediction in porous materials such as MOFs without structural information, and the techniques here can be used to better predict and understand optimal materials for various applications including, carbon dioxide capture, gas storage and separations.
This research was conducted in collaboration with Professor Dae-Woon Lim at Kyoto University, Professor Myunghyun Paik at Seoul National University, Professor Minyoung Yoon at Gachon University, and Aadesh Harale at Saudi Arabian Oil Company. The research was published in the Proceedings of the National Academy of Sciences (PNAS) online on 10 July and the co-first authors were Ph. D. candidate WooSeok Jeong and Professor Dae-Woon Lim.
This research was funded by the Saudi Aramco-KAIST CO2 Management Center.
(Figure 1. Trends in structure - material property map and in collapsed structures)
(Figure 2. Transferability between the experimental results of collapsed MOFs and the simulation results of crystalline MOFs)
2017.07.26 View 8544 -
 Bio-based p-Xylene Oxidation into Terephthalic Acid by Engineered E.coli
KAIST researchers have established an efficient biocatalytic system to produce terephthalic acid (TPA) from p-xylene (pX). It will allow this industrially important bulk chemical to be made available in a more environmentally-friendly manner.
The research team developed metabolically engineered Escherichia coli (E.coli) to biologically transform pX into TPA, a chemical necessary in the manufacturing of polyethylene terephthalate (PET). This biocatalysis system represents a greener and more efficient alternative to the traditional chemical methods for TPA production. This research, headed by Distinguished Professor Sang Yup Lee, was published in Nature Communications on May 31.
The research team utilized a metabolic engineering and synthetic biology approach to develop a recombinant microorganism that can oxidize pX into TPA using microbial fermentation. TPA is a globally important chemical commodity for manufacturing PET. It can be applied to manufacture plastic bottles, clothing fibers, films, and many other products. Currently, TPA is produced from pX oxidation through an industrially well-known chemical process (with a typical TPA yield of over 95 mol%), which shows, however, such drawbacks as intensive energy requirements at high temperatures and pressure, usage of heavy metal catalysts, and the unavoidable byproduct formation of 4-carboxybenzaldehyde.
The research team designed and constructed a synthetic metabolic pathway by incorporating the upper xylene degradation pathway of Pseudomonas putida F1 and the lower p-toluene sulfonate pathway of Comamonas testosteroni T-2, which successfully produced TPA from pX in small-scale cultures, with the formation of p-toluate (pTA) as the major byproduct. The team further optimized the pathway gene expression levels by using a synthetic biology toolkit, which gave the final engineered E. coli strain showing increased TPA production and the complete elimination of the byproduct.
Using this best-performing strain, the team designed an elegant two-phase (aqueous/organic) fermentation system for TPA production on a larger scale, where pX was supplied in the organic phase. Through a number of optimization steps, the team ultimately achieved production of 13.3 g TPA from 8.8 g pX, which represented an extraordinary yield of 97 mol%.
The team has developed a microbial biotechnology application which is reportedly the first successful example of the bio-based production of TPA from pX by the microbial fermentation of engineered E. coli. This bio-based TPA technology presents several advantages such as ambient reaction temperature and pressure, no use of heavy metals or other toxic chemicals, the removable of byproduct formation, and it is 100% environmentally compatible.
Professor Lee said, “We presented promising biotechnology for producing large amounts of the commodity chemical TPA, which is used for PET manufacturing, through metabolically engineered gut bacterium. Our research is meaningful in that it demonstrates the feasibility of the biotechnological production of bulk chemicals, and if reproducible when up-scaled, it will represent a breakthrough in hydrocarbon bioconversions.”
Ph.D. candidate Zi Wei Luo is the first author of this research (DOI:10.1038/ncomms15689).The research was supported by the Intelligent Synthetic Biology Center through the Global Frontier Project (2011-0031963) of the Ministry of Science, ICT & Future Planning through the National Research Foundation of Korea.
Figure: Biotransformation of pX into TPA by engineered E. coli.
This schematic diagram shows the overall conceptualization of how metabolically engineered E. coli produced TPA from pX. The engineered E. coli was developed through reconstituting a synthetic metabolic pathway for pX conversion to TPA and optimized for increased TPA yield and byproduct elimination. Two-phase partitioning fermentation system was developed for demonstrating the feasibility of large-scale production of TPA from pX using the engineered E. coli strains, where pX was supplied in the organic phase and TPA was produced in the aqueous phase.
2017.06.05 View 10397
Bio-based p-Xylene Oxidation into Terephthalic Acid by Engineered E.coli
KAIST researchers have established an efficient biocatalytic system to produce terephthalic acid (TPA) from p-xylene (pX). It will allow this industrially important bulk chemical to be made available in a more environmentally-friendly manner.
The research team developed metabolically engineered Escherichia coli (E.coli) to biologically transform pX into TPA, a chemical necessary in the manufacturing of polyethylene terephthalate (PET). This biocatalysis system represents a greener and more efficient alternative to the traditional chemical methods for TPA production. This research, headed by Distinguished Professor Sang Yup Lee, was published in Nature Communications on May 31.
The research team utilized a metabolic engineering and synthetic biology approach to develop a recombinant microorganism that can oxidize pX into TPA using microbial fermentation. TPA is a globally important chemical commodity for manufacturing PET. It can be applied to manufacture plastic bottles, clothing fibers, films, and many other products. Currently, TPA is produced from pX oxidation through an industrially well-known chemical process (with a typical TPA yield of over 95 mol%), which shows, however, such drawbacks as intensive energy requirements at high temperatures and pressure, usage of heavy metal catalysts, and the unavoidable byproduct formation of 4-carboxybenzaldehyde.
The research team designed and constructed a synthetic metabolic pathway by incorporating the upper xylene degradation pathway of Pseudomonas putida F1 and the lower p-toluene sulfonate pathway of Comamonas testosteroni T-2, which successfully produced TPA from pX in small-scale cultures, with the formation of p-toluate (pTA) as the major byproduct. The team further optimized the pathway gene expression levels by using a synthetic biology toolkit, which gave the final engineered E. coli strain showing increased TPA production and the complete elimination of the byproduct.
Using this best-performing strain, the team designed an elegant two-phase (aqueous/organic) fermentation system for TPA production on a larger scale, where pX was supplied in the organic phase. Through a number of optimization steps, the team ultimately achieved production of 13.3 g TPA from 8.8 g pX, which represented an extraordinary yield of 97 mol%.
The team has developed a microbial biotechnology application which is reportedly the first successful example of the bio-based production of TPA from pX by the microbial fermentation of engineered E. coli. This bio-based TPA technology presents several advantages such as ambient reaction temperature and pressure, no use of heavy metals or other toxic chemicals, the removable of byproduct formation, and it is 100% environmentally compatible.
Professor Lee said, “We presented promising biotechnology for producing large amounts of the commodity chemical TPA, which is used for PET manufacturing, through metabolically engineered gut bacterium. Our research is meaningful in that it demonstrates the feasibility of the biotechnological production of bulk chemicals, and if reproducible when up-scaled, it will represent a breakthrough in hydrocarbon bioconversions.”
Ph.D. candidate Zi Wei Luo is the first author of this research (DOI:10.1038/ncomms15689).The research was supported by the Intelligent Synthetic Biology Center through the Global Frontier Project (2011-0031963) of the Ministry of Science, ICT & Future Planning through the National Research Foundation of Korea.
Figure: Biotransformation of pX into TPA by engineered E. coli.
This schematic diagram shows the overall conceptualization of how metabolically engineered E. coli produced TPA from pX. The engineered E. coli was developed through reconstituting a synthetic metabolic pathway for pX conversion to TPA and optimized for increased TPA yield and byproduct elimination. Two-phase partitioning fermentation system was developed for demonstrating the feasibility of large-scale production of TPA from pX using the engineered E. coli strains, where pX was supplied in the organic phase and TPA was produced in the aqueous phase.
2017.06.05 View 10397 -
 Processable High Internal Phase Pickering Emulsion Using Depletion Attraction
Professor Siyoung Choi’s research team from the KAIST Department of Chemical & Biomolecular Engineering used physical force to successfully produce a stable emulsion.
Emulsions, commonly known as cosmetic products, refer to stably dispersed structures of oil droplets in water (or water droplets in oil). Pickering emulsions refer to emulsions stabilized using solid particles, instead of detergent. Traditionally, it is said that water and oil do not mix. Until recently, detergent was added to mix oil and water for dispersion. Emulsions have traditionally been produced using this technique and are currently used for products such as mayonnaise, sun block, and lotion.
On the other hand, Pickering emulsions have been used after stabilization of chemical treatments on solid particle surfaces to enhance adsorption power. However, there were limitations in its application, since the treatment process is complex and its applicable range remains limited. Instead of chemical treatment on Pickering emulsion surfaces, the research team mixed small macromolecules a few nanometer in size with larger solid particles (tens of nanometers to a few micrometers). This induced depletion force was used to successfully stabilize the emulsion.
Depletion force refers to the force a large number of small particles induces to aggregate the bigger particles, in order to secure free space for themselves. In short, the force induces an attraction between larger particles. Until now, depletion force could only be applied to solids and solid particles. However, the research team used macromolecules and large particles such as solid particles and oil droplets to show the applicability of depletion force between solids and liquids. By introducing macromolecules that act as smaller particles, hydrophilic solid particles enhanced the adsorption of solid particles to the oil droplet surface, while preventing dissociation from the particle surface, resulting in the maintenance of a stable state.
The research team confirmed the possibility of the simple production of various porous macromolecular materials using stable Pickering emulsions. Such porous macromolecules are expected to be applicable in separation film, systems engineering, drug delivery, and sensors, given their large surface area.
Professor KyuHan Kim, the first author said, “Until now, depletion force has only been used between solid colloid particles. This research has scientific significance since it is the first example of using depletion force between solid particles and liquid droplets.”
Professor Choi said, “Beyond its academic significance, this technology could contribute to industries and national competitiveness.” He continued, “Since this technology uses physical force, not chemical, to produce stable emulsion, it can be used regardless of the type of solid particle and macromolecule. Further, it could be used in customized porous material production for special purposes.”
The research was published in Nature Communications online on February 1. In particular, this research is significant since an undergraduate student, Subeen Kim, participated in the project as a second author through the KAIST Undergraduate Research Program (URP). This research was funded by the National Research Foundation of Korea.
(Figure 1: Images of the inner structure of porous macromolecules produced using the new technology)
(Figure 2: Images showing the measurement of rheological properties of Pickering emulsions and system processability)
(Figure 3: Images showing a stable Pickering emulsion system)
2017.04.19 View 8202
Processable High Internal Phase Pickering Emulsion Using Depletion Attraction
Professor Siyoung Choi’s research team from the KAIST Department of Chemical & Biomolecular Engineering used physical force to successfully produce a stable emulsion.
Emulsions, commonly known as cosmetic products, refer to stably dispersed structures of oil droplets in water (or water droplets in oil). Pickering emulsions refer to emulsions stabilized using solid particles, instead of detergent. Traditionally, it is said that water and oil do not mix. Until recently, detergent was added to mix oil and water for dispersion. Emulsions have traditionally been produced using this technique and are currently used for products such as mayonnaise, sun block, and lotion.
On the other hand, Pickering emulsions have been used after stabilization of chemical treatments on solid particle surfaces to enhance adsorption power. However, there were limitations in its application, since the treatment process is complex and its applicable range remains limited. Instead of chemical treatment on Pickering emulsion surfaces, the research team mixed small macromolecules a few nanometer in size with larger solid particles (tens of nanometers to a few micrometers). This induced depletion force was used to successfully stabilize the emulsion.
Depletion force refers to the force a large number of small particles induces to aggregate the bigger particles, in order to secure free space for themselves. In short, the force induces an attraction between larger particles. Until now, depletion force could only be applied to solids and solid particles. However, the research team used macromolecules and large particles such as solid particles and oil droplets to show the applicability of depletion force between solids and liquids. By introducing macromolecules that act as smaller particles, hydrophilic solid particles enhanced the adsorption of solid particles to the oil droplet surface, while preventing dissociation from the particle surface, resulting in the maintenance of a stable state.
The research team confirmed the possibility of the simple production of various porous macromolecular materials using stable Pickering emulsions. Such porous macromolecules are expected to be applicable in separation film, systems engineering, drug delivery, and sensors, given their large surface area.
Professor KyuHan Kim, the first author said, “Until now, depletion force has only been used between solid colloid particles. This research has scientific significance since it is the first example of using depletion force between solid particles and liquid droplets.”
Professor Choi said, “Beyond its academic significance, this technology could contribute to industries and national competitiveness.” He continued, “Since this technology uses physical force, not chemical, to produce stable emulsion, it can be used regardless of the type of solid particle and macromolecule. Further, it could be used in customized porous material production for special purposes.”
The research was published in Nature Communications online on February 1. In particular, this research is significant since an undergraduate student, Subeen Kim, participated in the project as a second author through the KAIST Undergraduate Research Program (URP). This research was funded by the National Research Foundation of Korea.
(Figure 1: Images of the inner structure of porous macromolecules produced using the new technology)
(Figure 2: Images showing the measurement of rheological properties of Pickering emulsions and system processability)
(Figure 3: Images showing a stable Pickering emulsion system)
2017.04.19 View 8202 -
 Nuclease-Resistant Hybrid Nanoflowers
An eco-friendly method to synthesize DNA-copper nanoflowers with high load efficiencies, low cytotoxicity, and strong resistance against nucleases has been developed by Professor Hyun Gyu Park in the Department of Chemical and Biomolecular Engineering and his collaborators.
The research team successfully formed a flower-shaped nanostructure in an eco-friendly condition by using interactions between copper ions and DNA containing amide and amine groups. The resulting nanoflowers exhibit high DNA loading capacities in addition to low cytotoxicity.
Flower-shaped nanocrystals called nanoflowers have gained attention for their distinct features of high surface roughness and high surface area to volume ratios. The nanoflowers have been used in many areas including catalysis, electronics, and analytical chemistry.
Of late, research breakthroughs were made in the generation of hybrid inorganic-organic nanoflowers containing various enzymes as organic components. The hybridization with inorganic materials greatly enhanced enzymatic activity, stability, and durability compared to the corresponding free enzymes.
Generally, the formation of protein nanocrystals requires high heat treatment so it has limitations for achieving the high loading capacities of intact DNA.
The research team addressed the issue, focusing on the fact that nucleic acids with well-defined structures and selective recognition properties also contain amide and amine groups in their nucleobases. They proved that flower-like structures could be formed by using nucleic acids as a synthetic template, which paved the way to synthesize the hybrid nanoflowers containing DNA as an organic component in an eco-friendly condition.
The team also confirmed that this synthetic method can be universally applied to any DNA sequences containing amide and amine groups. They said their approach is quite unique considering that the majority of previous works focused on the utilization of DNA as a linker to assemble the nanomaterials. They said the method has several advantageous features. First, the ‘green’ synthetic procedure doesn’t involve any toxic chemicals, and shows low cytotoxicity and strong resistance against nucleases. Second, the obtained nanoflowers exhibit exceptionally high DNA loading capacities.
Above all, such superior features of hybrid nanoflowers enabled the sensitive detection of various molecules including phenol, hydrogen peroxide, and glucose. DNA-copper nanoflowers showed even higher peroxidase activity than those of protein-copper nanoflowers, which may be due to the larger surface area of the flower- shaped structures, creating a greater chance for applying them in the field of sensing of detection of hydrogen peroxide.
The research team expects that their research will create diverse applications in many areas including biosensors and will be further applied into therapeutic applications.
Professor Park said, “The inorganic component in the hybrid nanoflowers not only exhibits low cytotoxicity, but also protects the encapsulated DNA from being cleaved by endonuclease enzymes. Using this feature, the nanostructure will be applied into developing gene therapeutic carriers.”
This research was co-led by Professor Moon Il Kim at Gachon University and KAIST graduate Ki Soo Park, currently a professor at Konkuk University, is the first author. The research was featured as the front cover article of the Journal of Materials Chemistry B on March 28, Issue 12, published by the Royal Society of Chemistry.
The research was funded by the Mid-Career Researcher Support Program of the National Research Foundation of Korea and the Global Frontier Project of the Ministry of Science, ICT & Future Planning.
(Figure: (A) Schematic illustration of the formation of nuclease-resistant DNA–inorganic nanoflowers. (B) SEM images showing time-dependent growth of DNA-nanoflowers. The concentration of A-rich ssDNA (Table S1, ESI†) was 0.25 mM.)
2017.04.14 View 8296
Nuclease-Resistant Hybrid Nanoflowers
An eco-friendly method to synthesize DNA-copper nanoflowers with high load efficiencies, low cytotoxicity, and strong resistance against nucleases has been developed by Professor Hyun Gyu Park in the Department of Chemical and Biomolecular Engineering and his collaborators.
The research team successfully formed a flower-shaped nanostructure in an eco-friendly condition by using interactions between copper ions and DNA containing amide and amine groups. The resulting nanoflowers exhibit high DNA loading capacities in addition to low cytotoxicity.
Flower-shaped nanocrystals called nanoflowers have gained attention for their distinct features of high surface roughness and high surface area to volume ratios. The nanoflowers have been used in many areas including catalysis, electronics, and analytical chemistry.
Of late, research breakthroughs were made in the generation of hybrid inorganic-organic nanoflowers containing various enzymes as organic components. The hybridization with inorganic materials greatly enhanced enzymatic activity, stability, and durability compared to the corresponding free enzymes.
Generally, the formation of protein nanocrystals requires high heat treatment so it has limitations for achieving the high loading capacities of intact DNA.
The research team addressed the issue, focusing on the fact that nucleic acids with well-defined structures and selective recognition properties also contain amide and amine groups in their nucleobases. They proved that flower-like structures could be formed by using nucleic acids as a synthetic template, which paved the way to synthesize the hybrid nanoflowers containing DNA as an organic component in an eco-friendly condition.
The team also confirmed that this synthetic method can be universally applied to any DNA sequences containing amide and amine groups. They said their approach is quite unique considering that the majority of previous works focused on the utilization of DNA as a linker to assemble the nanomaterials. They said the method has several advantageous features. First, the ‘green’ synthetic procedure doesn’t involve any toxic chemicals, and shows low cytotoxicity and strong resistance against nucleases. Second, the obtained nanoflowers exhibit exceptionally high DNA loading capacities.
Above all, such superior features of hybrid nanoflowers enabled the sensitive detection of various molecules including phenol, hydrogen peroxide, and glucose. DNA-copper nanoflowers showed even higher peroxidase activity than those of protein-copper nanoflowers, which may be due to the larger surface area of the flower- shaped structures, creating a greater chance for applying them in the field of sensing of detection of hydrogen peroxide.
The research team expects that their research will create diverse applications in many areas including biosensors and will be further applied into therapeutic applications.
Professor Park said, “The inorganic component in the hybrid nanoflowers not only exhibits low cytotoxicity, but also protects the encapsulated DNA from being cleaved by endonuclease enzymes. Using this feature, the nanostructure will be applied into developing gene therapeutic carriers.”
This research was co-led by Professor Moon Il Kim at Gachon University and KAIST graduate Ki Soo Park, currently a professor at Konkuk University, is the first author. The research was featured as the front cover article of the Journal of Materials Chemistry B on March 28, Issue 12, published by the Royal Society of Chemistry.
The research was funded by the Mid-Career Researcher Support Program of the National Research Foundation of Korea and the Global Frontier Project of the Ministry of Science, ICT & Future Planning.
(Figure: (A) Schematic illustration of the formation of nuclease-resistant DNA–inorganic nanoflowers. (B) SEM images showing time-dependent growth of DNA-nanoflowers. The concentration of A-rich ssDNA (Table S1, ESI†) was 0.25 mM.)
2017.04.14 View 8296