NAS
-
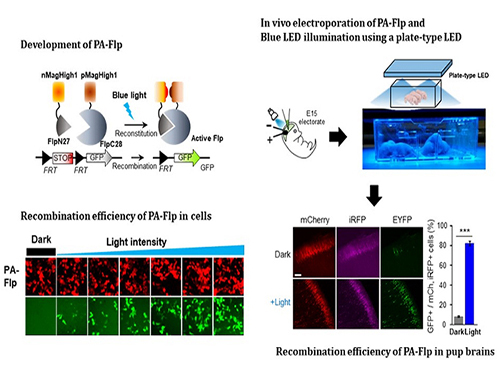 Noninvasive Light-Sensitive Recombinase for Deep Brain Genetic Manipulation
A KAIST team presented a noninvasive light-sensitive photoactivatable recombinase suitable for genetic manipulation in vivo. The highly light-sensitive property of photoactivatable Flp recombinase will be ideal for controlling genetic manipulation in deep mouse brain regions by illumination with a noninvasive light-emitting diode. This easy-to-use optogenetic module made by Professor Won Do Heo and his team will provide a side-effect free and expandable genetic manipulation tool for neuroscience research.
Spatiotemporal control of gene expression has been acclaimed as a valuable strategy for identifying functions of genes with complex neural circuits. Studies of complex brain functions require highly sophisticated and robust technologies that enable specific labeling and rapid genetic modification in live animals. A number of approaches for controlling the activity of proteins or expression of genes in a spatiotemporal manner using light, small molecules, hormones, and peptides have been developed for manipulating intact circuits or functions.
Among them, recombination-employing, chemically inducible systems are the most commonly used in vivo gene-modification systems. Other approaches include selective or conditional Cre-activation systems within subsets of green fluorescent protein-expressing cells or dual-promoter-driven intersectional populations of cells.
However, these methods are limited by the considerable time and effort required to establish knock-in mouse lines and by constraints on spatiotemporal control, which relies on a limited set of available genetic promoters and transgenic mouse resources.
Beyond these constraints, optogenetic approaches allow the activity of genetically defined neurons in the mouse brain to be controlled with high spatiotemporal resolution. However, an optogenetic module for gene-manipulation capable of revealing the spatiotemporal functions of specific target genes in the mouse brain has remained a challenge.
In the study published at Nature Communication on Jan. 18, the team featured photoactivatable Flp recombinase by searching out split sites of Flp recombinase that were not previously identified, being capable of reconstitution to be active. The team validated the highly light-sensitive, efficient performance of photoactivatable Flp recombinase through precise light targeting by showing transgene expression within anatomically confined mouse brain regions.
The concept of local genetic labeling presented here suggests a new approach for genetically identifying subpopulations of cells defined by the spatial and temporal characteristics of light delivery. To date, an optogenetic module for gene-manipulation capable of revealing spatiotemporal functions of specific target genes in the mouse brain has remained out of reach and no such light-inducible Flp system has been developed. Accordingly, the team sought to develop a photoactivatable Flp recombinase that takes full advantage of the high spatiotemporal control offered by light stimulation.
This activation through noninvasive light illumination deep inside the brain is advantageous in that it avoids chemical or optic fiber implantation-mediated side effects, such as off-target cytotoxicity or physical lesions that might influence animal physiology or behaviors. The technique provides expandable utilities for transgene expression systems upon Flp recombinase activity in vivo, by designing a viral vector for minimal leaky expression influenced by viral nascent promoters.
The team demonstrated the utility of PA-Flp as a noninvasive in vivo optogenetic manipulation tool for use in the mouse brain, even applicable for deep brain structures as it can reach the hippocampus or medial septum using external LED light illumination.
The study is the result of five years of research by Professor Heo, who has led the bio-imaging and optogenetics fields by developing his own bio-imaging and optogenetics technologies. “It will be a great advantage to control specific gene expression desired by LEDs with little physical and chemical stimulation that can affect the physiological phenomenon in living animals,” he explained.
2019.01.22 View 7944
Noninvasive Light-Sensitive Recombinase for Deep Brain Genetic Manipulation
A KAIST team presented a noninvasive light-sensitive photoactivatable recombinase suitable for genetic manipulation in vivo. The highly light-sensitive property of photoactivatable Flp recombinase will be ideal for controlling genetic manipulation in deep mouse brain regions by illumination with a noninvasive light-emitting diode. This easy-to-use optogenetic module made by Professor Won Do Heo and his team will provide a side-effect free and expandable genetic manipulation tool for neuroscience research.
Spatiotemporal control of gene expression has been acclaimed as a valuable strategy for identifying functions of genes with complex neural circuits. Studies of complex brain functions require highly sophisticated and robust technologies that enable specific labeling and rapid genetic modification in live animals. A number of approaches for controlling the activity of proteins or expression of genes in a spatiotemporal manner using light, small molecules, hormones, and peptides have been developed for manipulating intact circuits or functions.
Among them, recombination-employing, chemically inducible systems are the most commonly used in vivo gene-modification systems. Other approaches include selective or conditional Cre-activation systems within subsets of green fluorescent protein-expressing cells or dual-promoter-driven intersectional populations of cells.
However, these methods are limited by the considerable time and effort required to establish knock-in mouse lines and by constraints on spatiotemporal control, which relies on a limited set of available genetic promoters and transgenic mouse resources.
Beyond these constraints, optogenetic approaches allow the activity of genetically defined neurons in the mouse brain to be controlled with high spatiotemporal resolution. However, an optogenetic module for gene-manipulation capable of revealing the spatiotemporal functions of specific target genes in the mouse brain has remained a challenge.
In the study published at Nature Communication on Jan. 18, the team featured photoactivatable Flp recombinase by searching out split sites of Flp recombinase that were not previously identified, being capable of reconstitution to be active. The team validated the highly light-sensitive, efficient performance of photoactivatable Flp recombinase through precise light targeting by showing transgene expression within anatomically confined mouse brain regions.
The concept of local genetic labeling presented here suggests a new approach for genetically identifying subpopulations of cells defined by the spatial and temporal characteristics of light delivery. To date, an optogenetic module for gene-manipulation capable of revealing spatiotemporal functions of specific target genes in the mouse brain has remained out of reach and no such light-inducible Flp system has been developed. Accordingly, the team sought to develop a photoactivatable Flp recombinase that takes full advantage of the high spatiotemporal control offered by light stimulation.
This activation through noninvasive light illumination deep inside the brain is advantageous in that it avoids chemical or optic fiber implantation-mediated side effects, such as off-target cytotoxicity or physical lesions that might influence animal physiology or behaviors. The technique provides expandable utilities for transgene expression systems upon Flp recombinase activity in vivo, by designing a viral vector for minimal leaky expression influenced by viral nascent promoters.
The team demonstrated the utility of PA-Flp as a noninvasive in vivo optogenetic manipulation tool for use in the mouse brain, even applicable for deep brain structures as it can reach the hippocampus or medial septum using external LED light illumination.
The study is the result of five years of research by Professor Heo, who has led the bio-imaging and optogenetics fields by developing his own bio-imaging and optogenetics technologies. “It will be a great advantage to control specific gene expression desired by LEDs with little physical and chemical stimulation that can affect the physiological phenomenon in living animals,” he explained.
2019.01.22 View 7944 -
 Recombinant E. Coli As a Biofactory for the Biosynthesis of Diverse Nanomaterials
(Distinguished Professor Lee and PhD candidate Choi)
A metabolic research group at KAIST and Chung-Ang University in Korea has developed a recombinant E. coli strain that biosynthesizes 60 different nanomaterials covering 35 elements on the periodic table. Among the elements, the team could biosynthesize 33 novel nanomaterials for the first time, advancing the forward design of nanomaterials through the biosynthesis of various single and multi-elements.
The study analyzed the nanomaterial biosynthesis conditions using a Pourbaix diagram to predict the producibility and crystallinity. Researchers studied a Pourbaix diagram to predict the stable chemical species of each element for nanomaterial biosynthesis at varying levels of reduction potential (Eh) and pH. Based on the Pourbaix diagram analyses, the initial pH of the reaction was changed from 6.5 to 7.5, resulting in the biosynthesis of various crystalline nanomaterials that were previously amorphous or not synthesized.
This strategy was extended to biosynthesize multi-element nanomaterials. Various single and multi-element nanomaterials biosynthesized in this research can potentially serve as new and novel nanomaterials for industrial applications such as catalysts, chemical sensors, biosensors, bioimaging, drug delivery, and cancer therapy.
A research group consisting of PhD candidate Yoojin Choi, Associate Professor Doh Chang Lee, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST and Associate Professor Tae Jung Park of the Department of Chemistry at Chung-Ang University reported the synthesis. This study, entitled “Recombinant Escherichia coli as a biofactory for various single- and multi-element nanomaterials,” was published online in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on May 21.
A recent successful biosynthesis of nanomaterials under mild conditions without requiring physical and chemical treatments has triggered the exploration of the full biosynthesis capacity of a biological system for producing a diverse range of nanomaterials as well as for understanding biosynthesis mechanisms for crystalline versus amorphous nanomaterials.
There has been increased interest in synthesizing various nanomaterials that have not yet been synthesized for various applications including semiconducting materials, enhanced solar cells, biomedical materials, and many others. This research reports the construction of a recombinant E. coli strain that co-expresses metallothionein, a metal binding protein, and phytochelatin synthase that synthesizes the metal-binding peptide phytochelatin for the biosynthesis of various nanomaterials. Subsequently, an E. coli strain was engineered to produce a diverse range of nanomaterials, including those never biosynthesized before, by using 35 individual elements from the periodic table and also by combining multi-elements.
Distinguished Professor Lee said, “An environmentally-friendly and sustainable process is of much interest for producing nanomaterials by not only chemical and physical methods but biological synthesis. Moreover, there has been much attention paid to producing diverse and novel nanomaterials for new industrial applications. This is the first report to predict the biosynthesis of various nanomaterials, by far the largest number of various single- and multi-elements nanomaterials. The strategies used for nanomaterial biosynthesis in this research will be useful for further diversifying the portfolio of nanomaterials that can be manufactured.”
Figure: The biosynthesis of diverse nanomaterials using recombinant E. coli. This schematic diagram shows the overall conceptualization of the biosynthesis of various single and multi-element nanomaterials using recombinant E. coli under incubation with corresponding elemental precursors. The 35 elements that were tested to biosynthesize nanomaterials are shown in black circles on the periodic table.
2018.05.23 View 12418
Recombinant E. Coli As a Biofactory for the Biosynthesis of Diverse Nanomaterials
(Distinguished Professor Lee and PhD candidate Choi)
A metabolic research group at KAIST and Chung-Ang University in Korea has developed a recombinant E. coli strain that biosynthesizes 60 different nanomaterials covering 35 elements on the periodic table. Among the elements, the team could biosynthesize 33 novel nanomaterials for the first time, advancing the forward design of nanomaterials through the biosynthesis of various single and multi-elements.
The study analyzed the nanomaterial biosynthesis conditions using a Pourbaix diagram to predict the producibility and crystallinity. Researchers studied a Pourbaix diagram to predict the stable chemical species of each element for nanomaterial biosynthesis at varying levels of reduction potential (Eh) and pH. Based on the Pourbaix diagram analyses, the initial pH of the reaction was changed from 6.5 to 7.5, resulting in the biosynthesis of various crystalline nanomaterials that were previously amorphous or not synthesized.
This strategy was extended to biosynthesize multi-element nanomaterials. Various single and multi-element nanomaterials biosynthesized in this research can potentially serve as new and novel nanomaterials for industrial applications such as catalysts, chemical sensors, biosensors, bioimaging, drug delivery, and cancer therapy.
A research group consisting of PhD candidate Yoojin Choi, Associate Professor Doh Chang Lee, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST and Associate Professor Tae Jung Park of the Department of Chemistry at Chung-Ang University reported the synthesis. This study, entitled “Recombinant Escherichia coli as a biofactory for various single- and multi-element nanomaterials,” was published online in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on May 21.
A recent successful biosynthesis of nanomaterials under mild conditions without requiring physical and chemical treatments has triggered the exploration of the full biosynthesis capacity of a biological system for producing a diverse range of nanomaterials as well as for understanding biosynthesis mechanisms for crystalline versus amorphous nanomaterials.
There has been increased interest in synthesizing various nanomaterials that have not yet been synthesized for various applications including semiconducting materials, enhanced solar cells, biomedical materials, and many others. This research reports the construction of a recombinant E. coli strain that co-expresses metallothionein, a metal binding protein, and phytochelatin synthase that synthesizes the metal-binding peptide phytochelatin for the biosynthesis of various nanomaterials. Subsequently, an E. coli strain was engineered to produce a diverse range of nanomaterials, including those never biosynthesized before, by using 35 individual elements from the periodic table and also by combining multi-elements.
Distinguished Professor Lee said, “An environmentally-friendly and sustainable process is of much interest for producing nanomaterials by not only chemical and physical methods but biological synthesis. Moreover, there has been much attention paid to producing diverse and novel nanomaterials for new industrial applications. This is the first report to predict the biosynthesis of various nanomaterials, by far the largest number of various single- and multi-elements nanomaterials. The strategies used for nanomaterial biosynthesis in this research will be useful for further diversifying the portfolio of nanomaterials that can be manufactured.”
Figure: The biosynthesis of diverse nanomaterials using recombinant E. coli. This schematic diagram shows the overall conceptualization of the biosynthesis of various single and multi-element nanomaterials using recombinant E. coli under incubation with corresponding elemental precursors. The 35 elements that were tested to biosynthesize nanomaterials are shown in black circles on the periodic table.
2018.05.23 View 12418 -
 Professor Jun Ho Oh's Total Solar Eclipse Featured in the APOD, NASA
(Professor Jun Ho Oh)
A video of a total solar eclipse, filmed in Warm Springs, Oregon by Professor Jun Ho Oh of the Department of Mechanical Engineering, was selected as the Astronomy Picture of the Day (APOD).
APOD, is a NASA website specializing in astronomy pictures. It features astronomical observations recorded by the Hubble Space Telescope or photos taken by astronomical observers from around the world.
Professor Oh is now the second Korean and the first amateur photographer whose photo was selected as the APOD.
According to the website, ‘the video frames were acquired with equipment specifically designed by Jun Ho Oh to track a close-up of the Sun’s periphery during the eclipse.’
Also, Digital Photography Review (dpreview.com) introduced observation points of the eclipse in his three-minute video, including solar prominences, corona, and Baily’s beads.
Professor Oh, the creator of the bipedal walking humanoid robot named Hubo, has been chasing eclipse since his first trip to Turkey in 1999.
“After numerous trials and failures over the last 18 years, I was finally able to capture every single breath-taking moment of the total eclipse,” said the professor.
He’s already planning for the next total eclipse in Chile on July 2, 2019.
Click the link to watch the video https://apod.nasa.gov/apod/ap170912.html
(#1 Photo of solar eclipse)
(#2 Photo of solar eclipse)
2017.09.14 View 7683
Professor Jun Ho Oh's Total Solar Eclipse Featured in the APOD, NASA
(Professor Jun Ho Oh)
A video of a total solar eclipse, filmed in Warm Springs, Oregon by Professor Jun Ho Oh of the Department of Mechanical Engineering, was selected as the Astronomy Picture of the Day (APOD).
APOD, is a NASA website specializing in astronomy pictures. It features astronomical observations recorded by the Hubble Space Telescope or photos taken by astronomical observers from around the world.
Professor Oh is now the second Korean and the first amateur photographer whose photo was selected as the APOD.
According to the website, ‘the video frames were acquired with equipment specifically designed by Jun Ho Oh to track a close-up of the Sun’s periphery during the eclipse.’
Also, Digital Photography Review (dpreview.com) introduced observation points of the eclipse in his three-minute video, including solar prominences, corona, and Baily’s beads.
Professor Oh, the creator of the bipedal walking humanoid robot named Hubo, has been chasing eclipse since his first trip to Turkey in 1999.
“After numerous trials and failures over the last 18 years, I was finally able to capture every single breath-taking moment of the total eclipse,” said the professor.
He’s already planning for the next total eclipse in Chile on July 2, 2019.
Click the link to watch the video https://apod.nasa.gov/apod/ap170912.html
(#1 Photo of solar eclipse)
(#2 Photo of solar eclipse)
2017.09.14 View 7683 -
 Winning Best in Theme Award in NASA RASC-AL
A students team from the Department of Aerospace Engineering won the Best in Theme Award for moon exploration system design at Revolutionary Aerospace Systems Concepts - Academic Linkage (RASC-AL), an aerospace mission system design competition organized by NASA in the USA.
The KAIST team, consisting of Jaeyoul Ko, Jongeun Suh, Juseong Lee, Sukmin Choi, and Eunkwang Lee, and supervised by Professor Jaemyung Ahn, competed as a joint team with Texas Tech University and the Royal Melbourne Institute of Technology in Australia,
The joint team was selected as one of the 14 finalists after two preliminary rounds. The finals of RASC-AL Forum took place from May 30 to June 3 in Florida. The team received the top prize with their design entitled ‘Earth to Lunar Interchangeable Transportation Environment (ELITE) for Logistics Delivery Systems’, one of the four themes of the competition.
Since 2002, RASC-AL competitions, managed by NASA, have been held with themes on innovative aerospace system and missions, in which world-class undergraduate and graduate students have participated.
This year’s themes were ▲ Lightweight Exercise Suite ▲ Airlock Design ▲ Commercially Enabled LEO/Mars Habitable Module and ▲ Logistics Delivery System.
Moon exploration requires a great deal of time and supplies. The KAIST team has been researching supply delivery systems in space for long-term manned moon exploration with their joint team for the last eight months. In particular, incidents can occur during the initial stages of long-term manned moon exploration missions that are unpredictable during system design and planning. Therefore, to cope with such unpredictability in the mission, the KAIST team deduced a system and an operational concept with increased flexibility to maximize the cost effectiveness of the supply transport.
The spacecraft was divided into propulsion and transport modules based on their functionalities, and can allow the flexibility by switching the transport module according to the demands of the moon base. The operational flexibility and cost effectiveness are further increased by introducing multiple departure orbits from the Earth (e.g. low Earth orbit vs. geosynchronous Earth orbit) enabled by utilization of various launch vehicles.
Professor Ahn, the advisor for the team, said, “I am proud of the students who collaborated with the international joint teams and achieved great result.” He continued, “I believe this to be the result of continuous efforts and initiatives of the department for system design-centered education. We will keep providing high-quality system design and education through various opportunities such as international cooperation in design education.”
(Photo caption: KAIST team of the Department of Aerospace Engineering poses after winning the Best in Theme Award in NASA's RASC-AL)
2017.06.22 View 10219
Winning Best in Theme Award in NASA RASC-AL
A students team from the Department of Aerospace Engineering won the Best in Theme Award for moon exploration system design at Revolutionary Aerospace Systems Concepts - Academic Linkage (RASC-AL), an aerospace mission system design competition organized by NASA in the USA.
The KAIST team, consisting of Jaeyoul Ko, Jongeun Suh, Juseong Lee, Sukmin Choi, and Eunkwang Lee, and supervised by Professor Jaemyung Ahn, competed as a joint team with Texas Tech University and the Royal Melbourne Institute of Technology in Australia,
The joint team was selected as one of the 14 finalists after two preliminary rounds. The finals of RASC-AL Forum took place from May 30 to June 3 in Florida. The team received the top prize with their design entitled ‘Earth to Lunar Interchangeable Transportation Environment (ELITE) for Logistics Delivery Systems’, one of the four themes of the competition.
Since 2002, RASC-AL competitions, managed by NASA, have been held with themes on innovative aerospace system and missions, in which world-class undergraduate and graduate students have participated.
This year’s themes were ▲ Lightweight Exercise Suite ▲ Airlock Design ▲ Commercially Enabled LEO/Mars Habitable Module and ▲ Logistics Delivery System.
Moon exploration requires a great deal of time and supplies. The KAIST team has been researching supply delivery systems in space for long-term manned moon exploration with their joint team for the last eight months. In particular, incidents can occur during the initial stages of long-term manned moon exploration missions that are unpredictable during system design and planning. Therefore, to cope with such unpredictability in the mission, the KAIST team deduced a system and an operational concept with increased flexibility to maximize the cost effectiveness of the supply transport.
The spacecraft was divided into propulsion and transport modules based on their functionalities, and can allow the flexibility by switching the transport module according to the demands of the moon base. The operational flexibility and cost effectiveness are further increased by introducing multiple departure orbits from the Earth (e.g. low Earth orbit vs. geosynchronous Earth orbit) enabled by utilization of various launch vehicles.
Professor Ahn, the advisor for the team, said, “I am proud of the students who collaborated with the international joint teams and achieved great result.” He continued, “I believe this to be the result of continuous efforts and initiatives of the department for system design-centered education. We will keep providing high-quality system design and education through various opportunities such as international cooperation in design education.”
(Photo caption: KAIST team of the Department of Aerospace Engineering poses after winning the Best in Theme Award in NASA's RASC-AL)
2017.06.22 View 10219 -
 Distinguished Professor Lee Elected to the NAS
Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering was elected as a foreign associate to the US National Academy of Sciences (NAS) on May 2. The National Academy of Sciences elected 84 new members and 21 foreign associates in recognition of their distinguished and continuing achievements in their original research. Election to the Academy is widely regarded as one of the highest honors that a scientist can receive.
Professor Lee was also elected in 2010 as a member of the US National Academy of Engineering (NAE) for his leadership in microbial biotechnology and metabolic engineering, including the development of fermentation processes for biodegradable polymers and organic acids. Until 2016, there are only 12 people worldwide who are foreign associates of both NAS and NAE.
He is the first Korean elected to both prestigious academies, the NAS and the NAE in the US. Professor Lee is currently the dean of KAIST Institutes, the world leading institute for multi-and interdisciplinary research. He is also serving as co-chair of the Global Council on Biotechnology and member of the Global Future Council on the Fourth Industrial Revolution, the World Economic Forum.
2017.05.16 View 11126
Distinguished Professor Lee Elected to the NAS
Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering was elected as a foreign associate to the US National Academy of Sciences (NAS) on May 2. The National Academy of Sciences elected 84 new members and 21 foreign associates in recognition of their distinguished and continuing achievements in their original research. Election to the Academy is widely regarded as one of the highest honors that a scientist can receive.
Professor Lee was also elected in 2010 as a member of the US National Academy of Engineering (NAE) for his leadership in microbial biotechnology and metabolic engineering, including the development of fermentation processes for biodegradable polymers and organic acids. Until 2016, there are only 12 people worldwide who are foreign associates of both NAS and NAE.
He is the first Korean elected to both prestigious academies, the NAS and the NAE in the US. Professor Lee is currently the dean of KAIST Institutes, the world leading institute for multi-and interdisciplinary research. He is also serving as co-chair of the Global Council on Biotechnology and member of the Global Future Council on the Fourth Industrial Revolution, the World Economic Forum.
2017.05.16 View 11126 -
 Professor Seyun Kim Identifies a Neuron Signal Controlling Molecule
A research team led by Professor Seyun Kim of the Department of Biological Sciences at KAIST has identified inositol pyrophosphates as the molecule that strongly controls neuron signaling via synaptotagmin.
Professors Tae-Young Yoon of Yonsei University’s Y-IBS and Sung-Hyun Kim of Kyung Hee University’s Department of Biomedical Science also joined the team.
The results were published in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 30, 2016.
This interdisciplinary research project was conducted by six research teams from four different countries and covered a wide scope of academic fields, from neurobiology to super resolution optic imaging.
Inositol pyrophosphates such as 5-diphosphoinositol pentakisphos-phate (5-IP7), which naturally occur in corns and beans, are essential metabolites in the body. In particular, inositol hexakisphosphate (IP6) has anti-cancer properties and is thought to have an important role in cell signaling.
Inositol pentakisphosphate (IP7) differs from IP6 by having an additional phosphate group, which was first discovered 20 years ago. IP7 has recently been identified as playing a key role in diabetes and obesity.
Psychopathy and neurodegenerative diseases are known to result from the disrupted balance of inositol pyrophosphates. However, the role and the mechanism of action of IP7 in brain neurons and nerve transmission remained unknown.
Professor Kim’s team has worked on inositol pyrophosphates for several years and discovered that very small quantities of IP7 control cell-signaling transduction. Professor Yoon of Yonsei University identified IP7 as a much stronger inhibitor of neuron signaling compared to IP6. In particular, IP7 directly suppresses synaptotagmin, one of the key proteins in neuron signaling. Moreover, Professor Kim of Kyung Hee University observed IP7 inhibition in sea horse neurons.
Together, the joint research team identified inositol pyrophosphates as the key switch metabolite of brain-signaling transduction.
The researchers hope that future research on synaptotagmin and IP7 will reveal the mechanism of neuron-signal transduction and thus enable the treatment of neurological disorders.
These research findings were the result of cooperation of various science and technology institutes: KAIST, Yonsei-IBS (Institute for Basic Science), Kyung Hee University, Sungkyunkwan University, KIST, University of Zurich in Switzerland, and Albert-Ludwigs-University Freiburg in Germany.
Schematic Image of Controlling the Synaptic Exocytotic Pathway by 5-IP7 , Helping the Understanding of the Signaling Mechanisms of Inositol Pyrophosphates
2016.07.21 View 12852
Professor Seyun Kim Identifies a Neuron Signal Controlling Molecule
A research team led by Professor Seyun Kim of the Department of Biological Sciences at KAIST has identified inositol pyrophosphates as the molecule that strongly controls neuron signaling via synaptotagmin.
Professors Tae-Young Yoon of Yonsei University’s Y-IBS and Sung-Hyun Kim of Kyung Hee University’s Department of Biomedical Science also joined the team.
The results were published in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 30, 2016.
This interdisciplinary research project was conducted by six research teams from four different countries and covered a wide scope of academic fields, from neurobiology to super resolution optic imaging.
Inositol pyrophosphates such as 5-diphosphoinositol pentakisphos-phate (5-IP7), which naturally occur in corns and beans, are essential metabolites in the body. In particular, inositol hexakisphosphate (IP6) has anti-cancer properties and is thought to have an important role in cell signaling.
Inositol pentakisphosphate (IP7) differs from IP6 by having an additional phosphate group, which was first discovered 20 years ago. IP7 has recently been identified as playing a key role in diabetes and obesity.
Psychopathy and neurodegenerative diseases are known to result from the disrupted balance of inositol pyrophosphates. However, the role and the mechanism of action of IP7 in brain neurons and nerve transmission remained unknown.
Professor Kim’s team has worked on inositol pyrophosphates for several years and discovered that very small quantities of IP7 control cell-signaling transduction. Professor Yoon of Yonsei University identified IP7 as a much stronger inhibitor of neuron signaling compared to IP6. In particular, IP7 directly suppresses synaptotagmin, one of the key proteins in neuron signaling. Moreover, Professor Kim of Kyung Hee University observed IP7 inhibition in sea horse neurons.
Together, the joint research team identified inositol pyrophosphates as the key switch metabolite of brain-signaling transduction.
The researchers hope that future research on synaptotagmin and IP7 will reveal the mechanism of neuron-signal transduction and thus enable the treatment of neurological disorders.
These research findings were the result of cooperation of various science and technology institutes: KAIST, Yonsei-IBS (Institute for Basic Science), Kyung Hee University, Sungkyunkwan University, KIST, University of Zurich in Switzerland, and Albert-Ludwigs-University Freiburg in Germany.
Schematic Image of Controlling the Synaptic Exocytotic Pathway by 5-IP7 , Helping the Understanding of the Signaling Mechanisms of Inositol Pyrophosphates
2016.07.21 View 12852 -
 Public Lectures on the Korean Language and Alphabet
The School of Humanities and Social Sciences at KAIST will offer public lectures on the Korean language and alphabet, Hangul, from March 22, 2016 to April 26, 2016.
The lectures, which are entitled “The Riddle of Hangul,” will take place on campus in Daejeon.
A total of six lectures will be held on such topics as the origin of Korean, the grammar of ancient Korean in the Chosun Dynasty (1392-1897), and subsequent developments in contemporary Korean.
Professor Jung-Hoon Kim, who is responsible for organizing the public lecture program, said, “The audience will have an interesting opportunity to understand the history of Korean and its mechanism, while reviewing the unique spelling system of Hangul. I hope many people will show up for these wonderful classes.”
For further information and registration, please visit: http://hss.kaist.ac.kr. All lectures, available only in Korean, are free and open to the public.
2016.03.15 View 9990
Public Lectures on the Korean Language and Alphabet
The School of Humanities and Social Sciences at KAIST will offer public lectures on the Korean language and alphabet, Hangul, from March 22, 2016 to April 26, 2016.
The lectures, which are entitled “The Riddle of Hangul,” will take place on campus in Daejeon.
A total of six lectures will be held on such topics as the origin of Korean, the grammar of ancient Korean in the Chosun Dynasty (1392-1897), and subsequent developments in contemporary Korean.
Professor Jung-Hoon Kim, who is responsible for organizing the public lecture program, said, “The audience will have an interesting opportunity to understand the history of Korean and its mechanism, while reviewing the unique spelling system of Hangul. I hope many people will show up for these wonderful classes.”
For further information and registration, please visit: http://hss.kaist.ac.kr. All lectures, available only in Korean, are free and open to the public.
2016.03.15 View 9990 -
 Artificial Antibody-based Therapeutic Candidate for Lung Cancer Developed
Professor Hak-Sung Kim of Biological Sciences at KAIST publishes a cover article on artificial antibody in "Molecular Therapy".
Repebody-based lung cancer therapeutic drug candidate developed Repebody-based protein demonstrates the possibility of the development of a new drug
KAIST Biological Sciences Department’s Professor Hak-Sung Kim, in collaboration with Professor Eun-Kyung Cho from the College of Medicine at Chungnam National University, has successfully developed an artificial antibody-based, or repebody, cancer therapeutic candidate. These research results were published as a cover paper of the July edition of Molecular Therapy.
The repebody developed by Professor Kim and his team strongly binds to interleukin-6, a cancer-causing factor. It has also been confirmed that the repebody can significantly inhibit the proliferation of cancer cells in non-small-cell lung cancer animal model.
Numerous multinational pharmaceutical and biotechnology companies have invested astronomical amounts of money in research for the development of protein therapeutics with low side effects and high efficacy. More than 20 kinds of such therapeutics are currently under clinical trials, and over 100 drugs are under clinical demonstration. Among these, the majority is antibody-based therapeutics, and most of the investments are heavily concentrated in this field. However, antibody production cost is very high because it has large molecular weights and complex structural properties, and this makes it difficult to engineer. Consequently, the development costs a great deal of time and money.
In order to overcome the existing limitations of antibody-based therapeutics, Professor Kim and his team have developed a new artificial antibody, or repebody, which was published in Proceedings of the National Academy of Sciences (PNAS) in 2012. Based on this research, they have succeeded in developing a therapeutic candidate for treating non-small-cell lung cancer with a specifically strong cohesion to the cancer-causing factor, interleukin-6.
Interleukin-6 is a crucial substance within the body that is involved in immune and inflammatory-related signals. When abnormally expressed, it activates various carcinogenic pathways and promotes tumor growth and metastasis. Because of its importance, multinational pharmaceutical companies are heavily investing in developing therapeutics that can inhibit the signaling of interleukin-6.
In this study, Professor Kim and his team observed that a repebody consists of repeated modules, and they conceived a module-based affinity amplification technology that can effectively increase the binding affinity with the disease target. The developed therapeutic candidate has been confirmed in cell and animal experiments to show low immunogenicity, as well as to strongly inhibit the proliferation of non-small-cell lung cancer.
Furthermore, by investigating the complex structure of the repebody with interleukin-6, Professor Kim has identified its mechanism, which demonstrated the potential for therapeutic development. The researchers are currently carrying out pre-clinical trials for acquiring permission to perform clinical trials on animals with non-small-cell lung cancer. The repebody can be developed into a new protein drug after demonstrating its safety and efficacy.
Professor Hak-Sung Kim and his team have confirmed that the repebody can be utilized as a new protein drug, and this will be a significant contribution to Korea’s protein drugs and biotechnology industry development.
The research was supported by the Future Pioneer Industry project and sponsored by the Ministry of Science, ICT and Future Planning.
Figure 1. Professor Kim’s article published as the cover article of July edition of Molecular Therapy
Figure 2. Clinical proof of the repebody’s inhibition of cancer growth using animal models
2014.07.14 View 14339
Artificial Antibody-based Therapeutic Candidate for Lung Cancer Developed
Professor Hak-Sung Kim of Biological Sciences at KAIST publishes a cover article on artificial antibody in "Molecular Therapy".
Repebody-based lung cancer therapeutic drug candidate developed Repebody-based protein demonstrates the possibility of the development of a new drug
KAIST Biological Sciences Department’s Professor Hak-Sung Kim, in collaboration with Professor Eun-Kyung Cho from the College of Medicine at Chungnam National University, has successfully developed an artificial antibody-based, or repebody, cancer therapeutic candidate. These research results were published as a cover paper of the July edition of Molecular Therapy.
The repebody developed by Professor Kim and his team strongly binds to interleukin-6, a cancer-causing factor. It has also been confirmed that the repebody can significantly inhibit the proliferation of cancer cells in non-small-cell lung cancer animal model.
Numerous multinational pharmaceutical and biotechnology companies have invested astronomical amounts of money in research for the development of protein therapeutics with low side effects and high efficacy. More than 20 kinds of such therapeutics are currently under clinical trials, and over 100 drugs are under clinical demonstration. Among these, the majority is antibody-based therapeutics, and most of the investments are heavily concentrated in this field. However, antibody production cost is very high because it has large molecular weights and complex structural properties, and this makes it difficult to engineer. Consequently, the development costs a great deal of time and money.
In order to overcome the existing limitations of antibody-based therapeutics, Professor Kim and his team have developed a new artificial antibody, or repebody, which was published in Proceedings of the National Academy of Sciences (PNAS) in 2012. Based on this research, they have succeeded in developing a therapeutic candidate for treating non-small-cell lung cancer with a specifically strong cohesion to the cancer-causing factor, interleukin-6.
Interleukin-6 is a crucial substance within the body that is involved in immune and inflammatory-related signals. When abnormally expressed, it activates various carcinogenic pathways and promotes tumor growth and metastasis. Because of its importance, multinational pharmaceutical companies are heavily investing in developing therapeutics that can inhibit the signaling of interleukin-6.
In this study, Professor Kim and his team observed that a repebody consists of repeated modules, and they conceived a module-based affinity amplification technology that can effectively increase the binding affinity with the disease target. The developed therapeutic candidate has been confirmed in cell and animal experiments to show low immunogenicity, as well as to strongly inhibit the proliferation of non-small-cell lung cancer.
Furthermore, by investigating the complex structure of the repebody with interleukin-6, Professor Kim has identified its mechanism, which demonstrated the potential for therapeutic development. The researchers are currently carrying out pre-clinical trials for acquiring permission to perform clinical trials on animals with non-small-cell lung cancer. The repebody can be developed into a new protein drug after demonstrating its safety and efficacy.
Professor Hak-Sung Kim and his team have confirmed that the repebody can be utilized as a new protein drug, and this will be a significant contribution to Korea’s protein drugs and biotechnology industry development.
The research was supported by the Future Pioneer Industry project and sponsored by the Ministry of Science, ICT and Future Planning.
Figure 1. Professor Kim’s article published as the cover article of July edition of Molecular Therapy
Figure 2. Clinical proof of the repebody’s inhibition of cancer growth using animal models
2014.07.14 View 14339 -
 A game enthusiast received a Ph.D. at the 2014 commencement
A high school student, who was addicted to video gaming and had barely managed to gain entrance to KAIST, became a star of its 2014 commencement ceremony.
The student was Tae-Woo Park who received his Ph.D. in games at 32 years of age.
Park entered KAIST in 2002 as an undergraduate student. However, owning to bad grades, he was not accepted to the graduate school of KAIST until 2006. He began playing games at the age of 7, which distracted him from his studies at an early age. Nevertheless, he was able to complete master’s degree after two and a half years, which normally takes two years for average students.
Professor Joon-Hwa Song saw a possibility from his student’s experience of producing and commercializing a mobile puzzle game while Park was working as a president of the game club, HAJE, at KAIST. Professor Song advised him to take the advantage of his interests and try developing game platforms and contents. Park decided to develop a game that could help others and would change people’s negative views of games. He created a whole new generation of games.
In order to find ideas for games that can be easily enjoyed in daily lives, Park went to numerous gyms, swimming pools, daycare centers, and parks to analyze people’s behaviors and discussed with his colleagues who were also interested in games. During this process, the experience of organizing creative ideas through cooperation and discussions became a great foundation for his future research.
He observed some people quitting midway during a workout on treadmills because they were bored with working out alone. From this, Park embarked on developing a new style of game that allowed people to exercise together.
Park used the system on a treadmill, which recognizes the speed of the person running to automatically adjust the machine’s speed, to develop an interactive game platform for Swan Boat. The Swan Boat game is a race exercise game that adjusts the direction according to speed difference between two players. The game utilizes the difference of running speed between two people on treadmills to change the direction of the boat.
With the Swan Boat game, people can now play games and exercise at the same time. The technology also allows online access anywhere in the world, which means checking friends’ rankings at nearby gyms or homes, or even a World Gym Running Contest.
In addition, Park helped develop various next generation exercise games and life-based services, including the sparrow chirp application, which finds children that go astray, or an avatar game that utilizes the user’s daily life patterns. These results and papers attracted attention from international societies and have also won a number of awards.
Professor Song said, “There has been no precedent of receiving a Ph.D. at KAIST for developing games, however, Park’s case has given courage to many people that if you can create what is really required in everyday life, you can indeed receive a doctor’s degree.”
Park remarked, “I’d like to express my gratitude to my advisor, Professor Song, for giving me courage. I want to continue to make games that can help people’s lives in the future.”
Park will continue his work at the NASA Ames Research Center this June.
2014.02.27 View 11780
A game enthusiast received a Ph.D. at the 2014 commencement
A high school student, who was addicted to video gaming and had barely managed to gain entrance to KAIST, became a star of its 2014 commencement ceremony.
The student was Tae-Woo Park who received his Ph.D. in games at 32 years of age.
Park entered KAIST in 2002 as an undergraduate student. However, owning to bad grades, he was not accepted to the graduate school of KAIST until 2006. He began playing games at the age of 7, which distracted him from his studies at an early age. Nevertheless, he was able to complete master’s degree after two and a half years, which normally takes two years for average students.
Professor Joon-Hwa Song saw a possibility from his student’s experience of producing and commercializing a mobile puzzle game while Park was working as a president of the game club, HAJE, at KAIST. Professor Song advised him to take the advantage of his interests and try developing game platforms and contents. Park decided to develop a game that could help others and would change people’s negative views of games. He created a whole new generation of games.
In order to find ideas for games that can be easily enjoyed in daily lives, Park went to numerous gyms, swimming pools, daycare centers, and parks to analyze people’s behaviors and discussed with his colleagues who were also interested in games. During this process, the experience of organizing creative ideas through cooperation and discussions became a great foundation for his future research.
He observed some people quitting midway during a workout on treadmills because they were bored with working out alone. From this, Park embarked on developing a new style of game that allowed people to exercise together.
Park used the system on a treadmill, which recognizes the speed of the person running to automatically adjust the machine’s speed, to develop an interactive game platform for Swan Boat. The Swan Boat game is a race exercise game that adjusts the direction according to speed difference between two players. The game utilizes the difference of running speed between two people on treadmills to change the direction of the boat.
With the Swan Boat game, people can now play games and exercise at the same time. The technology also allows online access anywhere in the world, which means checking friends’ rankings at nearby gyms or homes, or even a World Gym Running Contest.
In addition, Park helped develop various next generation exercise games and life-based services, including the sparrow chirp application, which finds children that go astray, or an avatar game that utilizes the user’s daily life patterns. These results and papers attracted attention from international societies and have also won a number of awards.
Professor Song said, “There has been no precedent of receiving a Ph.D. at KAIST for developing games, however, Park’s case has given courage to many people that if you can create what is really required in everyday life, you can indeed receive a doctor’s degree.”
Park remarked, “I’d like to express my gratitude to my advisor, Professor Song, for giving me courage. I want to continue to make games that can help people’s lives in the future.”
Park will continue his work at the NASA Ames Research Center this June.
2014.02.27 View 11780 -
 Materials Developed for Sodium Rechargeable Battery by EEWS
The research group of Professor William Goddard III, You-Sung Jung, and Jang-Wook Choi from the Graduate School of Energy, Environment, Water, and Sustainability (EEWS) at KAIST has developed a new sodium-ion rechargeable battery which operates at a high voltage, can be charged, and stably discharges over 10,000 cycles. The research results were published in the online version of the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on December 30, 2013.
Since the material costs of sodium rechargeable batteries is 30 to 40 times lower than lithium batteries, it has received attention as an energy saving tool for smart grids and as the next generation of lithium rechargeable batteries. Until now, sodium-ion rechargeable batteries have had issues with stability when charging and discharging. The research group developed a vanadium-based electrode to solve these problems.
The group said follow-up research will be continued to develop advanced technology on sodium rechargeable batteries as it is still currently in the beginning stages.
The research team: From left to right is Professors William Goddard, You-Sung Jung, and Jang-Wook Choi
2014.01.13 View 12478
Materials Developed for Sodium Rechargeable Battery by EEWS
The research group of Professor William Goddard III, You-Sung Jung, and Jang-Wook Choi from the Graduate School of Energy, Environment, Water, and Sustainability (EEWS) at KAIST has developed a new sodium-ion rechargeable battery which operates at a high voltage, can be charged, and stably discharges over 10,000 cycles. The research results were published in the online version of the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on December 30, 2013.
Since the material costs of sodium rechargeable batteries is 30 to 40 times lower than lithium batteries, it has received attention as an energy saving tool for smart grids and as the next generation of lithium rechargeable batteries. Until now, sodium-ion rechargeable batteries have had issues with stability when charging and discharging. The research group developed a vanadium-based electrode to solve these problems.
The group said follow-up research will be continued to develop advanced technology on sodium rechargeable batteries as it is still currently in the beginning stages.
The research team: From left to right is Professors William Goddard, You-Sung Jung, and Jang-Wook Choi
2014.01.13 View 12478 -
 A powerful strategy for developing microbial cell factories by employing synthetic small RNAs
The current systems for the production of chemicals, fuels and materials heavily rely on the use of fossil resources. Due to the increasing concerns on climate change and other environmental problems, however, there has been much interest in developing biorefineries for the production of such chemicals, fuels and materials from renewable resources. For the biorefineries to be competitive with the traditional fossil resource-based refineries, development of high performance microorganisms is the most important as it will affect the overall economics of the process most significantly. Metabolic engineering, which can be defined as purposeful modification of cellular metabolic and regulatory networks with an aim to improve the production of a desired product, has been successfully employed to improve the performance of the cell. However, it is not trivial to engineer the cellular metabolism and regulatory circuits in the cell due to their high complexity.
In metabolic engineering, it is important to find the genes that need to be amplified and attenuated in order to increase the product formation rate while minimizing the production of undesirable byproducts. Gene knock-out experiments are often performed to delete those metabolic fluxes that will consequently result in the increase of the desired product formation. However, gene knock-out experiments require much effort and time to perform, and are difficult to do for a large number of genes. Furthermore, the gene knock-out experiments performed in one strain cannot be transferred to another organism and thus the whole experimental process has to be repeated. This is a big problem in developing a high performance microbial cell factory because it is required to find the best platform strain among many different strains. Therefore, researchers have been eager to develop a strategy that allows rapid identification of multiple genes to be attenuated in multiple strains at the same time.
A Korean research team led by Distinguished Professor Sang Yup Lee at the Department of Chemical and Biomolecular Engineering from the Korea Advanced Institute of Science and Technology (KAIST) reported the development of a strategy for efficiently developing microbial cell factories by employing synthetic small RNAs (sRNAs). They first reported the development of such system in Nature Biotechnology last February. This strategy of employing synthetic sRNAs in metabolic engineering has been receiving great interest worldwide as it allows easy, rapid, high-throughput, tunable, and un-doable knock-down of multiple genes in multiple strains at the same time. The research team published a paper online on August 8 as a cover page (September issue) in Nature Protocols, describing the detailed strategy and protocol to employ synthetic sRNAs for metabolic engineering.
In this paper, researchers described the detailed step-by-step protocol for synthetic sRNA-based gene expression control, including the sRNA design principles. Tailor-made synthetic sRNAs can be easily manipulated by using conventional gene cloning method. The use of synthetic sRNAs for gene expression regulation provides several advantages such as portability, conditionality, and tunability in high-throughput experiments. Plasmid-based synthetic sRNA expression system does not leave any scar on the chromosome, and can be easily transferred to many other host strains to be examined. Thus, the construction of libraries and examination of different host strains are much easier than the conventional hard-coded gene manipulation systems. Also, the expression of genes can be conditionally repressed by controlling the production of synthetic sRNAs. Synthetic sRNAs possessing different repression efficiencies make it possible to finely tune the gene expression levels as well. Furthermore, synthetic sRNAs allow knock-down of the expression of essential genes, which was not possible by conventional gene knock-out experiments.
Synthetic sRNAs can be utilized for diverse experiments where gene expression regulation is needed. One of promising applications is high-throughput screening of the target genes to be manipulated and multiple strains simultaneously to enhance the production of chemicals and materials of interest. Such simultaneous optimization of gene targets and strains has been one of the big challenges in metabolic engineering. Another application is to fine tune the expression of the screened genes for flux optimization, which would enhance chemical production further by balancing the flux between biomass formation and target chemical production. Synthetic sRNAs can also be applied to finely regulating genetic interactions in a circuit or network, which is essential in synthetic biology. Once a sRNA scaffold-harboring plasmid is constructed, tailor-made, synthetic sRNAs can be made within 3-4 days, followed by the desired application experiments.
Dr. Eytan Zlotorynski, an editor at Nature Protocols, said "This paper describes the detailed protocol for the design and applications of synthetic sRNA. The method, which has many advantages, is likely to become common practice, and prove useful for metabolic engineering and synthetic biology studies."
This paper published in Nature Protocols will be useful for all researchers in academia and industry who are interested in the use of synthetic sRNAs for fundamental and applied biological and biotechnological studies.
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012-C1AAA001-2012M1A2A2026556) and the Intelligent Synthetic Biology Center through the Global Frontier Project (2011-0031963) of the Ministry of Science, ICT and Future Planning through the National Research Foundation of Korea.
2013.10.31 View 10950
A powerful strategy for developing microbial cell factories by employing synthetic small RNAs
The current systems for the production of chemicals, fuels and materials heavily rely on the use of fossil resources. Due to the increasing concerns on climate change and other environmental problems, however, there has been much interest in developing biorefineries for the production of such chemicals, fuels and materials from renewable resources. For the biorefineries to be competitive with the traditional fossil resource-based refineries, development of high performance microorganisms is the most important as it will affect the overall economics of the process most significantly. Metabolic engineering, which can be defined as purposeful modification of cellular metabolic and regulatory networks with an aim to improve the production of a desired product, has been successfully employed to improve the performance of the cell. However, it is not trivial to engineer the cellular metabolism and regulatory circuits in the cell due to their high complexity.
In metabolic engineering, it is important to find the genes that need to be amplified and attenuated in order to increase the product formation rate while minimizing the production of undesirable byproducts. Gene knock-out experiments are often performed to delete those metabolic fluxes that will consequently result in the increase of the desired product formation. However, gene knock-out experiments require much effort and time to perform, and are difficult to do for a large number of genes. Furthermore, the gene knock-out experiments performed in one strain cannot be transferred to another organism and thus the whole experimental process has to be repeated. This is a big problem in developing a high performance microbial cell factory because it is required to find the best platform strain among many different strains. Therefore, researchers have been eager to develop a strategy that allows rapid identification of multiple genes to be attenuated in multiple strains at the same time.
A Korean research team led by Distinguished Professor Sang Yup Lee at the Department of Chemical and Biomolecular Engineering from the Korea Advanced Institute of Science and Technology (KAIST) reported the development of a strategy for efficiently developing microbial cell factories by employing synthetic small RNAs (sRNAs). They first reported the development of such system in Nature Biotechnology last February. This strategy of employing synthetic sRNAs in metabolic engineering has been receiving great interest worldwide as it allows easy, rapid, high-throughput, tunable, and un-doable knock-down of multiple genes in multiple strains at the same time. The research team published a paper online on August 8 as a cover page (September issue) in Nature Protocols, describing the detailed strategy and protocol to employ synthetic sRNAs for metabolic engineering.
In this paper, researchers described the detailed step-by-step protocol for synthetic sRNA-based gene expression control, including the sRNA design principles. Tailor-made synthetic sRNAs can be easily manipulated by using conventional gene cloning method. The use of synthetic sRNAs for gene expression regulation provides several advantages such as portability, conditionality, and tunability in high-throughput experiments. Plasmid-based synthetic sRNA expression system does not leave any scar on the chromosome, and can be easily transferred to many other host strains to be examined. Thus, the construction of libraries and examination of different host strains are much easier than the conventional hard-coded gene manipulation systems. Also, the expression of genes can be conditionally repressed by controlling the production of synthetic sRNAs. Synthetic sRNAs possessing different repression efficiencies make it possible to finely tune the gene expression levels as well. Furthermore, synthetic sRNAs allow knock-down of the expression of essential genes, which was not possible by conventional gene knock-out experiments.
Synthetic sRNAs can be utilized for diverse experiments where gene expression regulation is needed. One of promising applications is high-throughput screening of the target genes to be manipulated and multiple strains simultaneously to enhance the production of chemicals and materials of interest. Such simultaneous optimization of gene targets and strains has been one of the big challenges in metabolic engineering. Another application is to fine tune the expression of the screened genes for flux optimization, which would enhance chemical production further by balancing the flux between biomass formation and target chemical production. Synthetic sRNAs can also be applied to finely regulating genetic interactions in a circuit or network, which is essential in synthetic biology. Once a sRNA scaffold-harboring plasmid is constructed, tailor-made, synthetic sRNAs can be made within 3-4 days, followed by the desired application experiments.
Dr. Eytan Zlotorynski, an editor at Nature Protocols, said "This paper describes the detailed protocol for the design and applications of synthetic sRNA. The method, which has many advantages, is likely to become common practice, and prove useful for metabolic engineering and synthetic biology studies."
This paper published in Nature Protocols will be useful for all researchers in academia and industry who are interested in the use of synthetic sRNAs for fundamental and applied biological and biotechnological studies.
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012-C1AAA001-2012M1A2A2026556) and the Intelligent Synthetic Biology Center through the Global Frontier Project (2011-0031963) of the Ministry of Science, ICT and Future Planning through the National Research Foundation of Korea.
2013.10.31 View 10950 -
 Secondary, High Capacity Battery developed from Rice Husks
Rice husks, a waste product from rice polishing, has been successfully utilized as the silicon anode for use in high capacity lithium ion secondary batteries. The new silicon anode derived from rice husks exhibit superior output and lifespan.
Professor Choi Jang Wook (The Graduate School of Energy, Environment, Water and Sustainability (EEWS)) and Professor Park Seung Min (Department of Biochemistry) and their respective research teams separated naturally occurring, highly porous silica material within the rice husks and developed a 3-dimensional, highly porous silicon anode material.
The result of the research effort was published in the online edition of the Proceedings of the National Academy of Sciences (PNAS) journal, a world renowned journal in the field of natural sciences.
Silicon has attracted much attention as anode material for next generation lithium ion secondary batteries because it exhibits 3~5 times higher capacity than conventional graphene. The high capacity will pave the way to lithium secondary batteries with higher energy densities than conventional batteries. It is anticipated that the application of silicon batteries will yield electronic devices with a longer duration for use in addition to electronic vehicles boasting longer mileage.
The silicon anode is based on the 3-dimensional, highly porous structure of rice husks which remedies the problematic extreme volume expansion of conventional silicon anodes.
Utilization of inexpensive rice husks to create high value silicon anodes will cause a ripple effect on the industry and academia.
2013.08.23 View 11996
Secondary, High Capacity Battery developed from Rice Husks
Rice husks, a waste product from rice polishing, has been successfully utilized as the silicon anode for use in high capacity lithium ion secondary batteries. The new silicon anode derived from rice husks exhibit superior output and lifespan.
Professor Choi Jang Wook (The Graduate School of Energy, Environment, Water and Sustainability (EEWS)) and Professor Park Seung Min (Department of Biochemistry) and their respective research teams separated naturally occurring, highly porous silica material within the rice husks and developed a 3-dimensional, highly porous silicon anode material.
The result of the research effort was published in the online edition of the Proceedings of the National Academy of Sciences (PNAS) journal, a world renowned journal in the field of natural sciences.
Silicon has attracted much attention as anode material for next generation lithium ion secondary batteries because it exhibits 3~5 times higher capacity than conventional graphene. The high capacity will pave the way to lithium secondary batteries with higher energy densities than conventional batteries. It is anticipated that the application of silicon batteries will yield electronic devices with a longer duration for use in addition to electronic vehicles boasting longer mileage.
The silicon anode is based on the 3-dimensional, highly porous structure of rice husks which remedies the problematic extreme volume expansion of conventional silicon anodes.
Utilization of inexpensive rice husks to create high value silicon anodes will cause a ripple effect on the industry and academia.
2013.08.23 View 11996