GE
-
 Professor Alice Haeyun Oh to Join GPAI Expert Group
Professor Alice Haeyun Oh will participate in the Global Partnership on Artificial Intelligence (GPAI), an international and multi-stakeholder initiative hosted by the OECD to guide the responsible development and use of AI. In collaboration with partners and international organizations, GPAI will bring together leading experts from industry, civil society, government, and academia.
The Korean Ministry of Science and ICT (MSIT) officially announced that South Korea will take part in GPAI as one of the 15 founding members that include Canada, France, Japan, and the United States.
Professor Oh has been appointed as a new member of the Responsible AI Committee, one of the four committees that GPAI established along with the Data Governance Committee, Future of Work Committee, and Innovation and Commercialization Committee.
(END)
2020.06.22 View 9485
Professor Alice Haeyun Oh to Join GPAI Expert Group
Professor Alice Haeyun Oh will participate in the Global Partnership on Artificial Intelligence (GPAI), an international and multi-stakeholder initiative hosted by the OECD to guide the responsible development and use of AI. In collaboration with partners and international organizations, GPAI will bring together leading experts from industry, civil society, government, and academia.
The Korean Ministry of Science and ICT (MSIT) officially announced that South Korea will take part in GPAI as one of the 15 founding members that include Canada, France, Japan, and the United States.
Professor Oh has been appointed as a new member of the Responsible AI Committee, one of the four committees that GPAI established along with the Data Governance Committee, Future of Work Committee, and Innovation and Commercialization Committee.
(END)
2020.06.22 View 9485 -
 New Nanoparticle Drug Combination For Atherosclerosis
Physicochemical cargo-switching nanoparticles (CSNP) designed by KAIST can help significantly reduce cholesterol and macrophage foam cells in arteries, which are the two main triggers for atherosclerotic plaque and inflammation.
The CSNP-based combination drug delivery therapy was proved to exert cholesterol-lowering, anti-inflammatory, and anti-proliferative functions of two common medications for treating and preventing atherosclerosis that are cyclodextrin and statin. Professor Ji-Ho Park and Dr. Heegon Kim from KAIST’s Department of Bio and Brain Engineering said their study has shown great potential for future applications with reduced side effects.
Atherosclerosis is a chronic inflammatory vascular disease that is characterized by the accumulation of cholesterol and cholesterol-loaded macrophage foam cells in the intima. When this atherosclerotic plaque clogs and narrows the artery walls, they restrict blood flow and cause various cardiovascular conditions such as heart attacks and strokes. Heart attacks and strokes are the world’s first and fifth causes of death respectively.
Oral statin administration has been used in clinics as a standard care for atherosclerosis, which is prescribed to lower blood cholesterol and inhibit its accumulation within the plaque. Although statins can effectively prevent the progression of plaque growth, they have only shown modest efficacy in eliminating the already-established plaque. Therefore, patients are required to take statin drugs for the rest of their lives and will always carry the risk of plaque ruptures that can trigger a blood clot.
To address these issues, Professor Park and Dr. Kim exploited another antiatherogenic agent called cyclodextrin. In their paper published in the Journal of Controlled Release on March 10, Professor Park and Dr. Kim reported that the polymeric formulation of cyclodextrin with a diameter of approximately 10 nanometers(nm) can accumulate within the atherosclerotic plaque 14 times more and effectively reduce the plaque even at lower doses, compared to cyclodextrin in a non-polymer structure.
Moreover, although cyclodextrin is known to have a cytotoxic effect on hair cells in the cochlea, which can lead to hearing loss, cyclodextrin polymers developed by Professor Park’s research group exhibited a varying biodistribution profile and did not have this side effect.
In the follow-up study reported in ACS Nano on April 28, the researchers exploited both cyclodextrin and statin and form the cyclodextrin-statin self-assembly drug complex, based on previous findings that each drug can exert local anti-atherosclerosis effect within the plaque. The complex formation processes were optimized to obtain homogeneous and stable nanoparticles with a diameter of about 100 nm for systematic injection.
The therapeutic synergy of cyclodextrin and statin could reportedly enhance plaque-targeted drug delivery and anti-inflammation. Cyclodextrin led to the regression of cholesterol in the established plaque, and the statins were shown to inhibit the proliferation of macrophage foam cells. The study suggested that combination therapy is required to resolve the complex inflammatory cholesterol-rich microenvironment within the plaque.
Professor Park said, “While nanomedicine has been mainly developed for the treatment of cancers, our studies show that nanomedicine can also play a significant role in treating and preventing atherosclerosis, which causes various cardiovascular diseases that are the leading causes of death worldwide.”
This work was supported by KAIST and the National Research Foundation (NRF) of Korea.
Publications:
1. Heegon Kim, Junhee Han, and Ji-Ho Park. (2020) ‘Cyclodextrin polymer improves atherosclerosis therapy and reduces ototoxicity’ Journal of Controlled Release. Volume 319. Page 77-86. Available online at https://doi.org/10.1016/j.jconrel.2019.12.021
2. Kim, H., et al. (2020) ‘Affinity-Driven Design of Cargo-Switching Nanoparticles to Leverage a Cholesterol-Rich Microenvironment for Atherosclerosis Therapy’ ACS Nano. Available online at https://doi.org/10.1021/acsnano.9b08216
Profile: Ji-Ho Park, Ph.D.
Associate Professor
jihopark@kaist.ac.kr
http://openwetware.org/wiki/Park_Lab
Biomaterials Engineering Laboratory (BEL)
Department of Bio and Brain Engineering (BIOENG)
Korea Advanced Institute of Science and Technology (KAIST)
https://www.kaist.ac.kr
Daejeon 34141, Korea
Profile: Heegon Kim, Ph.D.
Postdoctoral Researcher
heegon@kaist.ac.kr
BEL, BIOENG, KAIST
(END)
2020.06.16 View 15542
New Nanoparticle Drug Combination For Atherosclerosis
Physicochemical cargo-switching nanoparticles (CSNP) designed by KAIST can help significantly reduce cholesterol and macrophage foam cells in arteries, which are the two main triggers for atherosclerotic plaque and inflammation.
The CSNP-based combination drug delivery therapy was proved to exert cholesterol-lowering, anti-inflammatory, and anti-proliferative functions of two common medications for treating and preventing atherosclerosis that are cyclodextrin and statin. Professor Ji-Ho Park and Dr. Heegon Kim from KAIST’s Department of Bio and Brain Engineering said their study has shown great potential for future applications with reduced side effects.
Atherosclerosis is a chronic inflammatory vascular disease that is characterized by the accumulation of cholesterol and cholesterol-loaded macrophage foam cells in the intima. When this atherosclerotic plaque clogs and narrows the artery walls, they restrict blood flow and cause various cardiovascular conditions such as heart attacks and strokes. Heart attacks and strokes are the world’s first and fifth causes of death respectively.
Oral statin administration has been used in clinics as a standard care for atherosclerosis, which is prescribed to lower blood cholesterol and inhibit its accumulation within the plaque. Although statins can effectively prevent the progression of plaque growth, they have only shown modest efficacy in eliminating the already-established plaque. Therefore, patients are required to take statin drugs for the rest of their lives and will always carry the risk of plaque ruptures that can trigger a blood clot.
To address these issues, Professor Park and Dr. Kim exploited another antiatherogenic agent called cyclodextrin. In their paper published in the Journal of Controlled Release on March 10, Professor Park and Dr. Kim reported that the polymeric formulation of cyclodextrin with a diameter of approximately 10 nanometers(nm) can accumulate within the atherosclerotic plaque 14 times more and effectively reduce the plaque even at lower doses, compared to cyclodextrin in a non-polymer structure.
Moreover, although cyclodextrin is known to have a cytotoxic effect on hair cells in the cochlea, which can lead to hearing loss, cyclodextrin polymers developed by Professor Park’s research group exhibited a varying biodistribution profile and did not have this side effect.
In the follow-up study reported in ACS Nano on April 28, the researchers exploited both cyclodextrin and statin and form the cyclodextrin-statin self-assembly drug complex, based on previous findings that each drug can exert local anti-atherosclerosis effect within the plaque. The complex formation processes were optimized to obtain homogeneous and stable nanoparticles with a diameter of about 100 nm for systematic injection.
The therapeutic synergy of cyclodextrin and statin could reportedly enhance plaque-targeted drug delivery and anti-inflammation. Cyclodextrin led to the regression of cholesterol in the established plaque, and the statins were shown to inhibit the proliferation of macrophage foam cells. The study suggested that combination therapy is required to resolve the complex inflammatory cholesterol-rich microenvironment within the plaque.
Professor Park said, “While nanomedicine has been mainly developed for the treatment of cancers, our studies show that nanomedicine can also play a significant role in treating and preventing atherosclerosis, which causes various cardiovascular diseases that are the leading causes of death worldwide.”
This work was supported by KAIST and the National Research Foundation (NRF) of Korea.
Publications:
1. Heegon Kim, Junhee Han, and Ji-Ho Park. (2020) ‘Cyclodextrin polymer improves atherosclerosis therapy and reduces ototoxicity’ Journal of Controlled Release. Volume 319. Page 77-86. Available online at https://doi.org/10.1016/j.jconrel.2019.12.021
2. Kim, H., et al. (2020) ‘Affinity-Driven Design of Cargo-Switching Nanoparticles to Leverage a Cholesterol-Rich Microenvironment for Atherosclerosis Therapy’ ACS Nano. Available online at https://doi.org/10.1021/acsnano.9b08216
Profile: Ji-Ho Park, Ph.D.
Associate Professor
jihopark@kaist.ac.kr
http://openwetware.org/wiki/Park_Lab
Biomaterials Engineering Laboratory (BEL)
Department of Bio and Brain Engineering (BIOENG)
Korea Advanced Institute of Science and Technology (KAIST)
https://www.kaist.ac.kr
Daejeon 34141, Korea
Profile: Heegon Kim, Ph.D.
Postdoctoral Researcher
heegon@kaist.ac.kr
BEL, BIOENG, KAIST
(END)
2020.06.16 View 15542 -
 Energy Storage Using Oxygen to Boost Battery Performance
Researchers have presented a novel electrode material for advanced energy storage device that is directly charged with oxygen from the air. Professor Jeung Ku Kang’s team synthesized and preserved the sub-nanometric particles of atomic cluster sizes at high mass loadings within metal-organic frameworks (MOF) by controlling the behavior of reactants at the molecular level. This new strategy ensures high performance for lithium-oxygen batteries, acclaimed as a next-generation energy storage technology and widely used in electric vehicles.
Lithium-oxygen batteries in principle can generate ten times higher energy densities than conventional lithium-ion batteries, but they suffer from very poor cyclability. One of the methods to improve cycle stability is to reduce the overpotential of electrocatalysts in cathode electrodes. When the size of an electrocatalyst material is reduced to the atomic level, the increased surface energy leads to increased activity while significantly accelerating the material’s agglomeration.
As a solution to this challenge, Professor Kang from the Department of Materials Science and Engineering aimed to maintain the improved activity by stabilizing atomic-scale sized electrocatalysts into the sub-nanometric spaces. This is a novel strategy for simultaneously producing and stabilizing atomic-level electrocatalysts within metal-organic frameworks (MOFs).
Metal-organic frameworks continuously assemble metal ions and organic linkers.
The team controlled hydrogen affinities between water molecules to separate them and transfer the isolated water molecules one by one through the sub-nanometric pores of MOFs. The transferred water molecules reacted with cobalt ions to form di-nuclear cobalt hydroxide under precisely controlled synthetic conditions, then the atomic-level cobalt hydroxide is stabilized inside the sub-nanometric pores.
The di-nuclear cobalt hydroxide that is stabilized in the sub-nanometric pores of metal-organic frameworks (MOFs) reduced the overpotential by 63.9% and showed ten-fold improvements in the life cycle.
Professor Kang said, “Simultaneously generating and stabilizing atomic-level electrocatalysts within MOFs can diversify materials according to numerous combinations of metal and organic linkers. It can expand not only the development of electrocatalysts, but also various research fields such as photocatalysts, medicine, the environment, and petrochemicals.”
This study was reported in Advanced Science (Title: Autogenous Production and Stabilization of Highly Loaded Sub-Nanometric Particles within Multishell Hollow Metal-Organic Frameworks and Their Utilization for High Performance in Li-O2 Batteries).
This research was mainly supported by the Global Frontier R&D Program of the Ministry of Science, ICT & Planning (Grant No. 2013M3A6B1078884) funded by the Ministry of Science, ICT & Future Planning, and the National Research Foundation of Korea (Grant No. 2019M3E6A1104196).
Profile:Professor Jeung Ku Kang
jeungku@kaist.ac.kr
http://nanosf.kaist.ac.kr/
Nano Materials Simulation and Fabrication Laboratory
Department of Materials Science and Engineering
KAIST
2020.06.15 View 14365
Energy Storage Using Oxygen to Boost Battery Performance
Researchers have presented a novel electrode material for advanced energy storage device that is directly charged with oxygen from the air. Professor Jeung Ku Kang’s team synthesized and preserved the sub-nanometric particles of atomic cluster sizes at high mass loadings within metal-organic frameworks (MOF) by controlling the behavior of reactants at the molecular level. This new strategy ensures high performance for lithium-oxygen batteries, acclaimed as a next-generation energy storage technology and widely used in electric vehicles.
Lithium-oxygen batteries in principle can generate ten times higher energy densities than conventional lithium-ion batteries, but they suffer from very poor cyclability. One of the methods to improve cycle stability is to reduce the overpotential of electrocatalysts in cathode electrodes. When the size of an electrocatalyst material is reduced to the atomic level, the increased surface energy leads to increased activity while significantly accelerating the material’s agglomeration.
As a solution to this challenge, Professor Kang from the Department of Materials Science and Engineering aimed to maintain the improved activity by stabilizing atomic-scale sized electrocatalysts into the sub-nanometric spaces. This is a novel strategy for simultaneously producing and stabilizing atomic-level electrocatalysts within metal-organic frameworks (MOFs).
Metal-organic frameworks continuously assemble metal ions and organic linkers.
The team controlled hydrogen affinities between water molecules to separate them and transfer the isolated water molecules one by one through the sub-nanometric pores of MOFs. The transferred water molecules reacted with cobalt ions to form di-nuclear cobalt hydroxide under precisely controlled synthetic conditions, then the atomic-level cobalt hydroxide is stabilized inside the sub-nanometric pores.
The di-nuclear cobalt hydroxide that is stabilized in the sub-nanometric pores of metal-organic frameworks (MOFs) reduced the overpotential by 63.9% and showed ten-fold improvements in the life cycle.
Professor Kang said, “Simultaneously generating and stabilizing atomic-level electrocatalysts within MOFs can diversify materials according to numerous combinations of metal and organic linkers. It can expand not only the development of electrocatalysts, but also various research fields such as photocatalysts, medicine, the environment, and petrochemicals.”
This study was reported in Advanced Science (Title: Autogenous Production and Stabilization of Highly Loaded Sub-Nanometric Particles within Multishell Hollow Metal-Organic Frameworks and Their Utilization for High Performance in Li-O2 Batteries).
This research was mainly supported by the Global Frontier R&D Program of the Ministry of Science, ICT & Planning (Grant No. 2013M3A6B1078884) funded by the Ministry of Science, ICT & Future Planning, and the National Research Foundation of Korea (Grant No. 2019M3E6A1104196).
Profile:Professor Jeung Ku Kang
jeungku@kaist.ac.kr
http://nanosf.kaist.ac.kr/
Nano Materials Simulation and Fabrication Laboratory
Department of Materials Science and Engineering
KAIST
2020.06.15 View 14365 -
 Unravelling Complex Brain Networks with Automated 3-D Neural Mapping
-Automated 3-D brain imaging data analysis technology offers more reliable and standardized analysis of the spatial organization of complex neural circuits.-
KAIST researchers developed a new algorithm for brain imaging data analysis that enables the precise and quantitative mapping of complex neural circuits onto a standardized 3-D reference atlas.
Brain imaging data analysis is indispensable in the studies of neuroscience. However, analysis of obtained brain imaging data has been heavily dependent on manual processing, which cannot guarantee the accuracy, consistency, and reliability of the results.
Conventional brain imaging data analysis typically begins with finding a 2-D brain atlas image that is visually similar to the experimentally obtained brain image. Then, the region-of-interest (ROI) of the atlas image is matched manually with the obtained image, and the number of labeled neurons in the ROI is counted.
Such a visual matching process between experimentally obtained brain images and 2-D brain atlas images has been one of the major sources of error in brain imaging data analysis, as the process is highly subjective, sample-specific, and susceptible to human error. Manual analysis processes for brain images are also laborious, and thus studying the complete 3-D neuronal organization on a whole-brain scale is a formidable task.
To address these issues, a KAIST research team led by Professor Se-Bum Paik from the Department of Bio and Brain Engineering developed new brain imaging data analysis software named 'AMaSiNe (Automated 3-D Mapping of Single Neurons)', and introduced the algorithm in the May 26 issue of Cell Reports.
AMaSiNe automatically detects the positions of single neurons from multiple brain images, and accurately maps all the data onto a common standard 3-D reference space. The algorithm allows the direct comparison of brain data from different animals by automatically matching similar features from the images, and computing the image similarity score.
This feature-based quantitative image-to-image comparison technology improves the accuracy, consistency, and reliability of analysis results using only a small number of brain slice image samples, and helps standardize brain imaging data analyses.
Unlike other existing brain imaging data analysis methods, AMaSiNe can also automatically find the alignment conditions from misaligned and distorted brain images, and draw an accurate ROI, without any cumbersome manual validation process.
AMaSiNe has been further proved to produce consistent results with brain slice images stained utilizing various methods including DAPI, Nissl, and autofluorescence.
The two co-lead authors of this study, Jun Ho Song and Woochul Choi, exploited these benefits of AMaSiNe to investigate the topographic organization of neurons that project to the primary visual area (VISp) in various ROIs, such as the dorsal lateral geniculate nucleus (LGd), which could hardly be addressed without proper calibration and standardization of the brain slice image samples.
In collaboration with Professor Seung-Hee Lee's group of the Department of Biological Science, the researchers successfully observed the 3-D topographic neural projections to the VISp from LGd, and also demonstrated that these projections could not be observed when the slicing angle was not properly corrected by AMaSiNe. The results suggest that the precise correction of a slicing angle is essential for the investigation of complex and important brain structures.
AMaSiNe is widely applicable in the studies of various brain regions and other experimental conditions. For example, in the research team’s previous study jointly conducted with Professor Yang Dan’s group at UC Berkeley, the algorithm enabled the accurate analysis of the neuronal subsets in the substantia nigra and their projections to the whole brain. Their findings were published in Science on January 24.
AMaSiNe is of great interest to many neuroscientists in Korea and abroad, and is being actively used by a number of other research groups at KAIST, MIT, Harvard, Caltech, and UC San Diego.
Professor Paik said, “Our new algorithm allows the spatial organization of complex neural circuits to be found in a standardized 3-D reference atlas on a whole-brain scale. This will bring brain imaging data analysis to a new level.”
He continued, “More in-depth insights for understanding the function of brain circuits can be achieved by facilitating more reliable and standardized analysis of the spatial organization of neural circuits in various regions of the brain.”
This work was supported by KAIST and the National Research Foundation of Korea (NRF).
Figure and Image Credit: Professor Se-Bum Paik, KAIST
Figure and Image Usage Restrictions: News organizations may use or redistribute these figures and images, with proper attribution, as part of news coverage of this paper only.
Publication:
Song, J. H., et al. (2020). Precise Mapping of Single Neurons by Calibrated 3D Reconstruction of Brain Slices Reveals Topographic Projection in Mouse Visual Cortex. Cell Reports. Volume 31, 107682. Available online at https://doi.org/10.1016/j.celrep.2020.107682
Profile:
Se-Bum Paik
Assistant Professor
sbpaik@kaist.ac.kr
http://vs.kaist.ac.kr/
VSNN Laboratory
Department of Bio and Brain Engineering
Program of Brain and Cognitive Engineering
http://kaist.ac.kr
Korea Advanced Institute of Science and Technology (KAIST)
Daejeon, Republic of Korea
(END)
2020.06.08 View 15303
Unravelling Complex Brain Networks with Automated 3-D Neural Mapping
-Automated 3-D brain imaging data analysis technology offers more reliable and standardized analysis of the spatial organization of complex neural circuits.-
KAIST researchers developed a new algorithm for brain imaging data analysis that enables the precise and quantitative mapping of complex neural circuits onto a standardized 3-D reference atlas.
Brain imaging data analysis is indispensable in the studies of neuroscience. However, analysis of obtained brain imaging data has been heavily dependent on manual processing, which cannot guarantee the accuracy, consistency, and reliability of the results.
Conventional brain imaging data analysis typically begins with finding a 2-D brain atlas image that is visually similar to the experimentally obtained brain image. Then, the region-of-interest (ROI) of the atlas image is matched manually with the obtained image, and the number of labeled neurons in the ROI is counted.
Such a visual matching process between experimentally obtained brain images and 2-D brain atlas images has been one of the major sources of error in brain imaging data analysis, as the process is highly subjective, sample-specific, and susceptible to human error. Manual analysis processes for brain images are also laborious, and thus studying the complete 3-D neuronal organization on a whole-brain scale is a formidable task.
To address these issues, a KAIST research team led by Professor Se-Bum Paik from the Department of Bio and Brain Engineering developed new brain imaging data analysis software named 'AMaSiNe (Automated 3-D Mapping of Single Neurons)', and introduced the algorithm in the May 26 issue of Cell Reports.
AMaSiNe automatically detects the positions of single neurons from multiple brain images, and accurately maps all the data onto a common standard 3-D reference space. The algorithm allows the direct comparison of brain data from different animals by automatically matching similar features from the images, and computing the image similarity score.
This feature-based quantitative image-to-image comparison technology improves the accuracy, consistency, and reliability of analysis results using only a small number of brain slice image samples, and helps standardize brain imaging data analyses.
Unlike other existing brain imaging data analysis methods, AMaSiNe can also automatically find the alignment conditions from misaligned and distorted brain images, and draw an accurate ROI, without any cumbersome manual validation process.
AMaSiNe has been further proved to produce consistent results with brain slice images stained utilizing various methods including DAPI, Nissl, and autofluorescence.
The two co-lead authors of this study, Jun Ho Song and Woochul Choi, exploited these benefits of AMaSiNe to investigate the topographic organization of neurons that project to the primary visual area (VISp) in various ROIs, such as the dorsal lateral geniculate nucleus (LGd), which could hardly be addressed without proper calibration and standardization of the brain slice image samples.
In collaboration with Professor Seung-Hee Lee's group of the Department of Biological Science, the researchers successfully observed the 3-D topographic neural projections to the VISp from LGd, and also demonstrated that these projections could not be observed when the slicing angle was not properly corrected by AMaSiNe. The results suggest that the precise correction of a slicing angle is essential for the investigation of complex and important brain structures.
AMaSiNe is widely applicable in the studies of various brain regions and other experimental conditions. For example, in the research team’s previous study jointly conducted with Professor Yang Dan’s group at UC Berkeley, the algorithm enabled the accurate analysis of the neuronal subsets in the substantia nigra and their projections to the whole brain. Their findings were published in Science on January 24.
AMaSiNe is of great interest to many neuroscientists in Korea and abroad, and is being actively used by a number of other research groups at KAIST, MIT, Harvard, Caltech, and UC San Diego.
Professor Paik said, “Our new algorithm allows the spatial organization of complex neural circuits to be found in a standardized 3-D reference atlas on a whole-brain scale. This will bring brain imaging data analysis to a new level.”
He continued, “More in-depth insights for understanding the function of brain circuits can be achieved by facilitating more reliable and standardized analysis of the spatial organization of neural circuits in various regions of the brain.”
This work was supported by KAIST and the National Research Foundation of Korea (NRF).
Figure and Image Credit: Professor Se-Bum Paik, KAIST
Figure and Image Usage Restrictions: News organizations may use or redistribute these figures and images, with proper attribution, as part of news coverage of this paper only.
Publication:
Song, J. H., et al. (2020). Precise Mapping of Single Neurons by Calibrated 3D Reconstruction of Brain Slices Reveals Topographic Projection in Mouse Visual Cortex. Cell Reports. Volume 31, 107682. Available online at https://doi.org/10.1016/j.celrep.2020.107682
Profile:
Se-Bum Paik
Assistant Professor
sbpaik@kaist.ac.kr
http://vs.kaist.ac.kr/
VSNN Laboratory
Department of Bio and Brain Engineering
Program of Brain and Cognitive Engineering
http://kaist.ac.kr
Korea Advanced Institute of Science and Technology (KAIST)
Daejeon, Republic of Korea
(END)
2020.06.08 View 15303 -
 The 10th KINC Fusion Research Awardees
The KAIST Institute for NanoCentury (KINC) recognized three distinguished researchers whose convergence studies made significant impacts. The KINC presented the 10th KINC Fusion Research Awards during a ceremony that took place at KAIST’s main campus in Daejeon on May 19.
This year’s ‘best’ convergence research award went to a joint research group led by Professor Hee Tak Kim from the Department of Chemical and Biomolecular Engineering and Professor Sang Ouk Kim from the Department of Materials Science and Engineering. Their research, featured in the December 27 issue of Advanced Materials as a front cover article last year, introduced the world’s first high-energy efficiency, membraneless, flowless, zinc-bromine battery. This study, in which research professor Gyoung Hwa Jeong, postdoctoral researcher Yearin Byun, and PhD candidate Ju-Hyuck Lee took part as co-lead authors, is deemed as an example of a best practice in convergence research in which two groups’ respective expertise in the fields of carbon materials and electrochemical analysis created a synergistic effect.
Professor Bumjoon Kim from the Department of Chemical and Biomolecular Engineering was also recognized for having published the most interdisciplinary research papers on polymer electronics and nanomaterials at home and abroad.
Professor Hee-Tae Jung, the Director of KINC and the host of the KINC Fusion Research Awards, said, “The KINC is happy to announce the 10th awardees in nano-fusion research this year. Since convergence is crucial for making revolutionary changes, the importance of convergence studies should be recognized. Our institute will spare no effort to create a research environment suitable for convergence studies, which will be crucial for making a significant difference.”
The KINC was established in June 2006 under the KAIST Institute with the mission of facilitating convergence studies by tearing down boarders among departments and carrying out interdisciplinary joint research. Currently, the institute is comprised of approximately 90 professors from 13 departments. It aims to become a hub of university institutes for nano-fusion research.
(END)
2020.05.19 View 16020
The 10th KINC Fusion Research Awardees
The KAIST Institute for NanoCentury (KINC) recognized three distinguished researchers whose convergence studies made significant impacts. The KINC presented the 10th KINC Fusion Research Awards during a ceremony that took place at KAIST’s main campus in Daejeon on May 19.
This year’s ‘best’ convergence research award went to a joint research group led by Professor Hee Tak Kim from the Department of Chemical and Biomolecular Engineering and Professor Sang Ouk Kim from the Department of Materials Science and Engineering. Their research, featured in the December 27 issue of Advanced Materials as a front cover article last year, introduced the world’s first high-energy efficiency, membraneless, flowless, zinc-bromine battery. This study, in which research professor Gyoung Hwa Jeong, postdoctoral researcher Yearin Byun, and PhD candidate Ju-Hyuck Lee took part as co-lead authors, is deemed as an example of a best practice in convergence research in which two groups’ respective expertise in the fields of carbon materials and electrochemical analysis created a synergistic effect.
Professor Bumjoon Kim from the Department of Chemical and Biomolecular Engineering was also recognized for having published the most interdisciplinary research papers on polymer electronics and nanomaterials at home and abroad.
Professor Hee-Tae Jung, the Director of KINC and the host of the KINC Fusion Research Awards, said, “The KINC is happy to announce the 10th awardees in nano-fusion research this year. Since convergence is crucial for making revolutionary changes, the importance of convergence studies should be recognized. Our institute will spare no effort to create a research environment suitable for convergence studies, which will be crucial for making a significant difference.”
The KINC was established in June 2006 under the KAIST Institute with the mission of facilitating convergence studies by tearing down boarders among departments and carrying out interdisciplinary joint research. Currently, the institute is comprised of approximately 90 professors from 13 departments. It aims to become a hub of university institutes for nano-fusion research.
(END)
2020.05.19 View 16020 -
 Highly Efficient Charge-to-Spin Interconversion in Graphene Heterostructures
Researchers present a new route for designing a graphene-based active spintronic component
KAIST physicists described a route to design the energy-efficient generation, manipulation and detection of spin currents using nonmagnetic two-dimensional materials. The research team, led by Professor Sungjae Cho, observed highly efficient charge-to-spin interconversion via the gate-tunable Rashba-Edelstien effect (REE) in graphene heterostructures.
This research paves the way for the application of graphene as an active spintronic component for generating, controlling, and detecting spin current without ferromagnetic electrodes or magnetic fields.
Graphene is a promising spintronic component owing to its long spin diffusion length. However, its small spin-orbit coupling limits the potential of graphene in spintronic applications since graphene cannot be used to generate, control, or detect spin current.
“We successfully increased the spin-orbit coupling of graphene by stacking graphene on top of 2H-TaS2, which is one of the transition metal dichalcogenide materials with the largest spin-orbit coupling. Graphene now can be used to generate, control, and detect spin current,” Professor Cho said.
The Rashba-Edelstein effect is a physical mechanism that enables charge current-to-spin current interconversion by spin-dependent band structure induced by the Rashba effect, a momentum-dependent splitting of spin bands in low-dimensional condensed matter systems.
Professor Cho’s group demonstrated the gate-tunable Rashba-Edelstein effect in a multilayer graphene for the first time. The Rahsba-Edelstein effect allows the two-dimensional conduction electrons of graphene to be magnetized by an applied charge current and form a spin current. Furthermore, as the Fermi level of graphene, tuned by gate voltage, moves from the valence to conduction band, the spin current generated by graphene reversed its spin direction.
This spin reversal is useful in the design of low-power-consumption transistors utilizing spins in that it provides the carrier “On” state with spin up holes (or spin down electrons) and the "Off" state with zero net spin polarization at so called “charge neutrality point” where numbers of electrons and holes are equal.
“Our work is the first demonstration of charge-to-spin interconversion in a metallic TMD (transition-metal dichalcogenides) and graphene heterostructure with a spin polarization state controlled by a gate. We expect that the all-electrical spin-switching effect and the reversal of non-equilibrium spin polarization by the application of gate voltage is applicable for the energy-efficient generation and manipulation of spin currents using nonmagnetic van der Waals materials,” explained Professor Cho.
This study (https://pubs.acs.org/doi/10.1021/acsnano.0c01037) was supported by the National Research Foundation of Korea.
Publication:
Lijun Li, Jin Zhang, Gyuho Myeong, Wongil Shin, Hongsik Lim, Boram Kim, Seungho Kim, Taehyeok Jin, Stuart Cavill, Beom Seo Kim, Changyoung Kim, Johannes Lischner, Aires Ferreira, and Sungjae Cho, Gate-Tunable Reversible Rashba−Edelstein Effect in a Few-Layer Graphene/2H-TaS2 Heterostructure at Room Temperature. ACS Nano 2020. Link to download the paper: https://pubs.acs.org/doi/10.1021/acsnano.0c01037
Profile:
Professor Sungjae Cho, PhD
sungjae.cho@kaist.ac.kr
http://qtak.kaist.ac.kr
Department of Physics
Korea Advanced Institute of Science and Technology (KAIST)
https://www.kaist.ac.kr
Daejeon 34141, Korea
2020.05.18 View 11317
Highly Efficient Charge-to-Spin Interconversion in Graphene Heterostructures
Researchers present a new route for designing a graphene-based active spintronic component
KAIST physicists described a route to design the energy-efficient generation, manipulation and detection of spin currents using nonmagnetic two-dimensional materials. The research team, led by Professor Sungjae Cho, observed highly efficient charge-to-spin interconversion via the gate-tunable Rashba-Edelstien effect (REE) in graphene heterostructures.
This research paves the way for the application of graphene as an active spintronic component for generating, controlling, and detecting spin current without ferromagnetic electrodes or magnetic fields.
Graphene is a promising spintronic component owing to its long spin diffusion length. However, its small spin-orbit coupling limits the potential of graphene in spintronic applications since graphene cannot be used to generate, control, or detect spin current.
“We successfully increased the spin-orbit coupling of graphene by stacking graphene on top of 2H-TaS2, which is one of the transition metal dichalcogenide materials with the largest spin-orbit coupling. Graphene now can be used to generate, control, and detect spin current,” Professor Cho said.
The Rashba-Edelstein effect is a physical mechanism that enables charge current-to-spin current interconversion by spin-dependent band structure induced by the Rashba effect, a momentum-dependent splitting of spin bands in low-dimensional condensed matter systems.
Professor Cho’s group demonstrated the gate-tunable Rashba-Edelstein effect in a multilayer graphene for the first time. The Rahsba-Edelstein effect allows the two-dimensional conduction electrons of graphene to be magnetized by an applied charge current and form a spin current. Furthermore, as the Fermi level of graphene, tuned by gate voltage, moves from the valence to conduction band, the spin current generated by graphene reversed its spin direction.
This spin reversal is useful in the design of low-power-consumption transistors utilizing spins in that it provides the carrier “On” state with spin up holes (or spin down electrons) and the "Off" state with zero net spin polarization at so called “charge neutrality point” where numbers of electrons and holes are equal.
“Our work is the first demonstration of charge-to-spin interconversion in a metallic TMD (transition-metal dichalcogenides) and graphene heterostructure with a spin polarization state controlled by a gate. We expect that the all-electrical spin-switching effect and the reversal of non-equilibrium spin polarization by the application of gate voltage is applicable for the energy-efficient generation and manipulation of spin currents using nonmagnetic van der Waals materials,” explained Professor Cho.
This study (https://pubs.acs.org/doi/10.1021/acsnano.0c01037) was supported by the National Research Foundation of Korea.
Publication:
Lijun Li, Jin Zhang, Gyuho Myeong, Wongil Shin, Hongsik Lim, Boram Kim, Seungho Kim, Taehyeok Jin, Stuart Cavill, Beom Seo Kim, Changyoung Kim, Johannes Lischner, Aires Ferreira, and Sungjae Cho, Gate-Tunable Reversible Rashba−Edelstein Effect in a Few-Layer Graphene/2H-TaS2 Heterostructure at Room Temperature. ACS Nano 2020. Link to download the paper: https://pubs.acs.org/doi/10.1021/acsnano.0c01037
Profile:
Professor Sungjae Cho, PhD
sungjae.cho@kaist.ac.kr
http://qtak.kaist.ac.kr
Department of Physics
Korea Advanced Institute of Science and Technology (KAIST)
https://www.kaist.ac.kr
Daejeon 34141, Korea
2020.05.18 View 11317 -
 Hubo Debuts as a News Anchor
HUBO, a humanoid robot developed by Professor Jun-Ho Oh’s team, made its debut as a co-anchor during the TJB prime time news 8 on May 14.
“Un-contact" became the new normal after Covid-19 and many business solutions are being transformed using robotics. HUBO made two news reports on contactless services using robots in medical, manufacturing, and logistics industries. HUBO 2, the second generation of HUBO, appeared as a special anchor on the local broadcasting network’s special program in celebration of its 25th anniversary.
HUBO is the champion of the 2015 DARPA Robotics Challenge held in the USA. Its FX-2 riding robot also participated in the Olympic torch relay during the 2018 PyeongChang Winter Olympics.
Click here to watch a full video of HUBO anchoring the news.
(END)
2020.05.14 View 13960
Hubo Debuts as a News Anchor
HUBO, a humanoid robot developed by Professor Jun-Ho Oh’s team, made its debut as a co-anchor during the TJB prime time news 8 on May 14.
“Un-contact" became the new normal after Covid-19 and many business solutions are being transformed using robotics. HUBO made two news reports on contactless services using robots in medical, manufacturing, and logistics industries. HUBO 2, the second generation of HUBO, appeared as a special anchor on the local broadcasting network’s special program in celebration of its 25th anniversary.
HUBO is the champion of the 2015 DARPA Robotics Challenge held in the USA. Its FX-2 riding robot also participated in the Olympic torch relay during the 2018 PyeongChang Winter Olympics.
Click here to watch a full video of HUBO anchoring the news.
(END)
2020.05.14 View 13960 -
 Simple Molecular Reagents to Treat Alzheimer’s Disease
- Researchers report minimalistic principles for designing small molecules with multiple reactivities against dementia. -
Sometimes the most complex problems actually have very simple solutions. A group of South Korean researchers reported an efficient and effective redox-based strategy for incorporating multiple functions into simple molecular reagents against neurodegenerative disorders. The team developed redox-active aromatic molecular reagents with a simple structural composition that can simultaneously target and modulate various pathogenic factors in complex neurodegenerative disorders such as Alzheimer’s disease.
Alzheimer’s disease is one of the most prevalent neurodegenerative disorders, affecting one in ten people over the age of 65. Early-onset dementia also increasingly affects younger people.
A number of pathogenic elements such as reactive oxygen species, amyloid-beta, and metal ions have been suggested as potential causes of Alzheimer’s disease. Each element itself can lead to Alzheimer’s disease, but interactions between them may also aggravate the patient’s condition or interfere with the appropriate clinical care.
For example, when interacting with amyloid-beta, metal ions foster the aggregation and accumulation of amyloid-beta peptides that can induce oxidative stress and toxicity in the brain and lead to neurodegeneration.
Because these pathogenic factors of Alzheimer’s disease are intertwined, developing therapeutic agents that are capable of simultaneously regulating metal ion dyshomeostasis, amyloid-beta agglutination, and oxidative stress responses remains a key to halting the progression of the disease.
A research team led by Professor Mi Hee Lim from the Department of Chemistry at KAIST demonstrated the feasibility of structure-mechanism-based molecular design for controlling a molecule’s chemical reactivity toward the various pathological factors of Alzheimer’s disease by tuning the redox properties of the molecule.
This study, featured as the ‘ACS Editors’ Choice’ in the May 6th issue of the Journal of the American Chemical Society (JACS), was conducted in conjunction with KAIST Professor Mu-Hyun Baik’s group and Professor Joo-Young Lee’s group at the Asan Medical Center.
Professor Lim and her collaborators rationally designed and generated 10 compact aromatic molecules presenting a range of redox potentials by adjusting the electronic distribution of the phenyl, phenylene, or pyridyl moiety to impart redox-dependent reactivities against the multiple pathogenic factors in Alzheimer’s disease.
During the team’s biochemical and biophysical studies, these designed molecular reagents displayed redox-dependent reactivities against numerous desirable targets that are associated with Alzheimer’s disease such as free radicals, metal-free amyloid-beta, and metal-bound amyloid-beta.
Further mechanistic results revealed that the redox properties of these designed molecular reagents were essential for their function. The team demonstrated that these reagents engaged in oxidative reactions with metal-free and metal-bound amyloid-beta and led to chemical modifications. The products of such oxidative transformations were observed to form covalent adducts with amyloid-beta and alter its aggregation.
Moreover, the administration of the most promising candidate molecule significantly attenuated the amyloid pathology in the brains of Alzheimer’s disease transgenic mice and improved their cognitive defects.
Professor Lim said, “This strategy is straightforward, time-saving, and cost-effective, and its effect is significant. We are excited to help enable the advancement of new therapeutic agents for neurodegenerative disorders, which can improve the lives of so many patients.”
This work was supported by the National Research Foundation (NRF) of Korea, the Institute for Basic Science (IBS), and the Asan Institute for Life Sciences.
Image credit: Professor Mi Hee Lim, KAIST
Image usage restrictions: News organizations may use or redistribute this image, with proper attribution, as part of the news coverage of this paper only.
Publication:
Kim, M. et al. (2020) ‘Minimalistic Principles for Designing Small Molecules with Multiple Reactivities against Pathological Factors in Dementia.’ Journal of the American Chemical Society (JACS), Volume 142, Issue 18, pp.8183-8193. Available online at https://doi.org/10.1021/jacs.9b13100
Profile:
Mi Hee Lim
Professor
miheelim@kaist.ac.kr
http://sites.google.com/site/miheelimlab
Lim Laboratory
Department of Chemistry
KAIST
Profile:
Mu-Hyun Baik
Professor
mbaik2805@kaist.ac.kr
https://baik-laboratory.com/
Baik Laboratory
Department of Chemistry
KAIST
Profile:
Joo-Yong Lee
Professor
jlee@amc.seoul.kr
Asan Institute for Life Sciences
Asan Medical Center
(END)
2020.05.11 View 17848
Simple Molecular Reagents to Treat Alzheimer’s Disease
- Researchers report minimalistic principles for designing small molecules with multiple reactivities against dementia. -
Sometimes the most complex problems actually have very simple solutions. A group of South Korean researchers reported an efficient and effective redox-based strategy for incorporating multiple functions into simple molecular reagents against neurodegenerative disorders. The team developed redox-active aromatic molecular reagents with a simple structural composition that can simultaneously target and modulate various pathogenic factors in complex neurodegenerative disorders such as Alzheimer’s disease.
Alzheimer’s disease is one of the most prevalent neurodegenerative disorders, affecting one in ten people over the age of 65. Early-onset dementia also increasingly affects younger people.
A number of pathogenic elements such as reactive oxygen species, amyloid-beta, and metal ions have been suggested as potential causes of Alzheimer’s disease. Each element itself can lead to Alzheimer’s disease, but interactions between them may also aggravate the patient’s condition or interfere with the appropriate clinical care.
For example, when interacting with amyloid-beta, metal ions foster the aggregation and accumulation of amyloid-beta peptides that can induce oxidative stress and toxicity in the brain and lead to neurodegeneration.
Because these pathogenic factors of Alzheimer’s disease are intertwined, developing therapeutic agents that are capable of simultaneously regulating metal ion dyshomeostasis, amyloid-beta agglutination, and oxidative stress responses remains a key to halting the progression of the disease.
A research team led by Professor Mi Hee Lim from the Department of Chemistry at KAIST demonstrated the feasibility of structure-mechanism-based molecular design for controlling a molecule’s chemical reactivity toward the various pathological factors of Alzheimer’s disease by tuning the redox properties of the molecule.
This study, featured as the ‘ACS Editors’ Choice’ in the May 6th issue of the Journal of the American Chemical Society (JACS), was conducted in conjunction with KAIST Professor Mu-Hyun Baik’s group and Professor Joo-Young Lee’s group at the Asan Medical Center.
Professor Lim and her collaborators rationally designed and generated 10 compact aromatic molecules presenting a range of redox potentials by adjusting the electronic distribution of the phenyl, phenylene, or pyridyl moiety to impart redox-dependent reactivities against the multiple pathogenic factors in Alzheimer’s disease.
During the team’s biochemical and biophysical studies, these designed molecular reagents displayed redox-dependent reactivities against numerous desirable targets that are associated with Alzheimer’s disease such as free radicals, metal-free amyloid-beta, and metal-bound amyloid-beta.
Further mechanistic results revealed that the redox properties of these designed molecular reagents were essential for their function. The team demonstrated that these reagents engaged in oxidative reactions with metal-free and metal-bound amyloid-beta and led to chemical modifications. The products of such oxidative transformations were observed to form covalent adducts with amyloid-beta and alter its aggregation.
Moreover, the administration of the most promising candidate molecule significantly attenuated the amyloid pathology in the brains of Alzheimer’s disease transgenic mice and improved their cognitive defects.
Professor Lim said, “This strategy is straightforward, time-saving, and cost-effective, and its effect is significant. We are excited to help enable the advancement of new therapeutic agents for neurodegenerative disorders, which can improve the lives of so many patients.”
This work was supported by the National Research Foundation (NRF) of Korea, the Institute for Basic Science (IBS), and the Asan Institute for Life Sciences.
Image credit: Professor Mi Hee Lim, KAIST
Image usage restrictions: News organizations may use or redistribute this image, with proper attribution, as part of the news coverage of this paper only.
Publication:
Kim, M. et al. (2020) ‘Minimalistic Principles for Designing Small Molecules with Multiple Reactivities against Pathological Factors in Dementia.’ Journal of the American Chemical Society (JACS), Volume 142, Issue 18, pp.8183-8193. Available online at https://doi.org/10.1021/jacs.9b13100
Profile:
Mi Hee Lim
Professor
miheelim@kaist.ac.kr
http://sites.google.com/site/miheelimlab
Lim Laboratory
Department of Chemistry
KAIST
Profile:
Mu-Hyun Baik
Professor
mbaik2805@kaist.ac.kr
https://baik-laboratory.com/
Baik Laboratory
Department of Chemistry
KAIST
Profile:
Joo-Yong Lee
Professor
jlee@amc.seoul.kr
Asan Institute for Life Sciences
Asan Medical Center
(END)
2020.05.11 View 17848 -
 Breastfeeding Helps Prevent Mothers from Developing Diabetes after Childbirth
A team of South Korean researchers found that lactation can lower the incidence and reduce the risk of maternal postpartum diabetes. The researchers identified that lactation increases the mass and function of pancreatic beta cells through serotonin production. The team suggested that sustained improvements in pancreatic beta cells, which can last for years even after the cessation of lactation, improve mothers’ metabolic health in addition to providing health benefits for infants.
Pregnancy imposes a substantial metabolic burden on women through weight gain and increased insulin resistance. Various other factors, including a history of gestational diabetes, maternal age, and obesity, further affect women’s risk of progressing to diabetes after delivery, and the risk of postpartum diabetes increases more in women who have had gestational diabetes and/or repeated deliveries.
Diabetes-related complications include damage to blood vessels, which can lead to cardiovascular and cerebrovascular diseases such as heart attack and stroke, and problems with the nerves, eyes, kidneys, and many more. Since diabetes can pose a serious threat to mothers’ metabolic health, the management of maternal metabolic risk factors is important, especially in the peripartum period. Previous epidemiological studies have reported that lactation reduces the risk of postpartum diabetes, but the mechanisms underlying this benefit have remained elusive.
The study, published in Science Translational Medicine on April 29, explains the biology underpinning this observation on the beneficial effects of lactation. Professor Hail Kim from the Graduate School of Medical Science and Engineering at KAIST led and jointly conducted the study in conjunction with researchers from the Seoul National University Bundang Hospital (SNUBH) and Chungnam National University (CNU) in Korea, and the University of California, San Francisco (UCSF) in the US.
In their study, the team observed that the milk-secreting hormone ‘prolactin’ in lactating mothers not only promotes milk production, but also plays a major role in stimulating insulin-secreting pancreatic beta cells that regulate blood glucose in the body.
The researchers also found that ‘serotonin’, known as a chemical that contributes to wellbeing and happiness, is produced in pancreatic beta cells during lactation. Serotonin in pancreatic beta cells act as an antioxidant and reduce oxidative stress, making mothers’ beta cells healthier. Serotonin also induces the proliferation of beta cells, thereby increasing the beta cell mass and helping maintain proper glucose levels.
The research team conducted follow-up examinations on a total of 174 postpartum women, 85 lactated and 99 non-lactated, at two months postpartum and annually thereafter for at least three years. The results demonstrated that mothers who had undergone lactation improved pancreatic beta cell mass and function, and showed improved glucose homeostasis with approximately 20mg/dL lower glucose levels, thereby reducing the risk of postpartum diabetes in women. Surprisingly, this beneficial effect was maintained after the cessation of lactation, for more than three years after delivery.
Professor Kim said, “We are happy to prove that lactation benefits female metabolic health by improving beta cell mass and function as well as glycemic control.”
“Our future studies on the modulation of the molecular serotonergic pathway in accordance with the management of maternal metabolic risk factors may lead to new therapeutics to help prevent mothers from developing metabolic disorders,” he added.
This work was supported by grants from the National Research Foundation (NRF) and the National Research Council of Science and Technology (NST) of Korea, the National Institutes of Health (NIH), the Larry L. Hillblom Foundation, and the Health Fellowship Foundation.
Image credit: Professor Hail Kim, KAIST
Image usage restrictions: News organizations may use or redistribute this image, with proper attribution, as part of news coverage of this paper only.
Publication:
Moon, J. H et al. (2020) ‘Lactation improves pancreatic β cell mass and function through serotonin production.’ Science Translational Medicine, 12, eaay0455. Available online at https://doi.org/10.1126/scitranslmed.aay0455
Profile: Hail Kim, MD, PhD
hailkim@kaist.edu
Associate Professor
Graduate School of Medical Science and Engineering (GSMSE)
Korea Advanced Institute of Science and Technology (KAIST)
Profile: Hak Chul Jang, MD, PhD
janghak@snu.ac.kr
Professor
Division of Endocrinology and Metabolism
Seoul National University Bundang Hospital (SNUBH)
President
Korean Diabetes Association
Profile: Joon Ho Moon, MD, PhD
moonjoonho@gmail.com
Clinical Fellow
Division of Endocrinology and Metabolism
SNUBH
Profile: Hyeongseok Kim, MD, PhD
hskim85kor@gmail.com
Assistant Professor
Chungnam National University (CNU)
Profile: Professor Michael S. German, MD
Michael.German@ucsf.edu
Professor
Diabetes Center
University of California, San Francisco (UCSF)
(END)
2020.04.29 View 22019
Breastfeeding Helps Prevent Mothers from Developing Diabetes after Childbirth
A team of South Korean researchers found that lactation can lower the incidence and reduce the risk of maternal postpartum diabetes. The researchers identified that lactation increases the mass and function of pancreatic beta cells through serotonin production. The team suggested that sustained improvements in pancreatic beta cells, which can last for years even after the cessation of lactation, improve mothers’ metabolic health in addition to providing health benefits for infants.
Pregnancy imposes a substantial metabolic burden on women through weight gain and increased insulin resistance. Various other factors, including a history of gestational diabetes, maternal age, and obesity, further affect women’s risk of progressing to diabetes after delivery, and the risk of postpartum diabetes increases more in women who have had gestational diabetes and/or repeated deliveries.
Diabetes-related complications include damage to blood vessels, which can lead to cardiovascular and cerebrovascular diseases such as heart attack and stroke, and problems with the nerves, eyes, kidneys, and many more. Since diabetes can pose a serious threat to mothers’ metabolic health, the management of maternal metabolic risk factors is important, especially in the peripartum period. Previous epidemiological studies have reported that lactation reduces the risk of postpartum diabetes, but the mechanisms underlying this benefit have remained elusive.
The study, published in Science Translational Medicine on April 29, explains the biology underpinning this observation on the beneficial effects of lactation. Professor Hail Kim from the Graduate School of Medical Science and Engineering at KAIST led and jointly conducted the study in conjunction with researchers from the Seoul National University Bundang Hospital (SNUBH) and Chungnam National University (CNU) in Korea, and the University of California, San Francisco (UCSF) in the US.
In their study, the team observed that the milk-secreting hormone ‘prolactin’ in lactating mothers not only promotes milk production, but also plays a major role in stimulating insulin-secreting pancreatic beta cells that regulate blood glucose in the body.
The researchers also found that ‘serotonin’, known as a chemical that contributes to wellbeing and happiness, is produced in pancreatic beta cells during lactation. Serotonin in pancreatic beta cells act as an antioxidant and reduce oxidative stress, making mothers’ beta cells healthier. Serotonin also induces the proliferation of beta cells, thereby increasing the beta cell mass and helping maintain proper glucose levels.
The research team conducted follow-up examinations on a total of 174 postpartum women, 85 lactated and 99 non-lactated, at two months postpartum and annually thereafter for at least three years. The results demonstrated that mothers who had undergone lactation improved pancreatic beta cell mass and function, and showed improved glucose homeostasis with approximately 20mg/dL lower glucose levels, thereby reducing the risk of postpartum diabetes in women. Surprisingly, this beneficial effect was maintained after the cessation of lactation, for more than three years after delivery.
Professor Kim said, “We are happy to prove that lactation benefits female metabolic health by improving beta cell mass and function as well as glycemic control.”
“Our future studies on the modulation of the molecular serotonergic pathway in accordance with the management of maternal metabolic risk factors may lead to new therapeutics to help prevent mothers from developing metabolic disorders,” he added.
This work was supported by grants from the National Research Foundation (NRF) and the National Research Council of Science and Technology (NST) of Korea, the National Institutes of Health (NIH), the Larry L. Hillblom Foundation, and the Health Fellowship Foundation.
Image credit: Professor Hail Kim, KAIST
Image usage restrictions: News organizations may use or redistribute this image, with proper attribution, as part of news coverage of this paper only.
Publication:
Moon, J. H et al. (2020) ‘Lactation improves pancreatic β cell mass and function through serotonin production.’ Science Translational Medicine, 12, eaay0455. Available online at https://doi.org/10.1126/scitranslmed.aay0455
Profile: Hail Kim, MD, PhD
hailkim@kaist.edu
Associate Professor
Graduate School of Medical Science and Engineering (GSMSE)
Korea Advanced Institute of Science and Technology (KAIST)
Profile: Hak Chul Jang, MD, PhD
janghak@snu.ac.kr
Professor
Division of Endocrinology and Metabolism
Seoul National University Bundang Hospital (SNUBH)
President
Korean Diabetes Association
Profile: Joon Ho Moon, MD, PhD
moonjoonho@gmail.com
Clinical Fellow
Division of Endocrinology and Metabolism
SNUBH
Profile: Hyeongseok Kim, MD, PhD
hskim85kor@gmail.com
Assistant Professor
Chungnam National University (CNU)
Profile: Professor Michael S. German, MD
Michael.German@ucsf.edu
Professor
Diabetes Center
University of California, San Francisco (UCSF)
(END)
2020.04.29 View 22019 -
 A Study Finds Neuropeptide Somatostatin Enhances Visual Processing
Researchers have confirmed that neuropeptide somatostatin can improve cognitive function in the brain. A research group of Professor Seung-Hee Lee from the Department of Biological Sciences at KAIST found that the application of neuropeptide somatostatin improves visual processing and cognitive behaviors by reducing excitatory inputs to parvalbumin-positive interneurons in the cortex.
This study, reported at Science Advances on April 22nd (EST), sheds a new light on the therapeutics of neurodegenerative diseases. According to a recent study in Korea, one in ten seniors over 65 is experiencing dementia-related symptoms in their daily lives such like memory loss, cognitive decline, and motion function disorders. Professor Lee believes that somatostatin treatment can be directly applied to the recovery of cognitive functions in Alzheimer’s disease patients.
Professor Lee started this study noting the fact that the level of somatostatin expression was dramatically decreased in the cerebral cortex and cerebrospinal fluid of Alzheimer’s disease patients
Somatostatin-expressing neurons in the cortex are known to exert the dendritic inhibition of pyramidal neurons via GABAergic transmission. Previous studies focused on their inhibitory effects on cortical circuits, but somatostatin-expressing neurons can co-release somatostatin upon activation. Despite the abundant expression of somatostatin and its receptors in the cerebral cortex, it was not known if somatostatin could modulate cognitive processing in the cortex.
The research team demonstrated that the somatostatin treatment into the cerebral cortex could enhance visual processing and cognitive behaviors in mice. The research team combined behaviors, in vivo and in vitro electrophysiology, and electron microscopy techniques to reveal how the activation of somatostatin receptors in vivo enhanced the ability of visual recognition in animals. Interestingly, somatostatin release can reduce excitatory synaptic transmission to another subtype of GABAergic interneurons, parvalbumin (PV)-expressing neurons.
As somatostatin is a stable and safe neuropeptide expressed naturally in the mammalian brain, it was safe to be injected into the cortex and cerebrospinal fluid, showing a potential application to drug development for curing cognitive disorders in humans.
Professor Lee said, “Our research confirmed the key role of the neuropeptide SST in modulating cortical function and enhancing cognitive ability in the mammalian brain. I hope new drugs can be developed based on the function of somatostatin to treat cognitive disabilities in many patients suffering from neurological disorders.”
This study was supported by the National Research Foundation of Korea.
Publication:
Song, Y. H et al. (2020) ‘Somatostatin enhances visual processing and perception by suppressing excitatory inputs to parvalbumin-positive interneurons in V1’, Science Advances, 6(17). Available online at https://doi.org/10.1126/sciadv.aaz0517
Profile:
Seung-Hee Lee
Associate Professor
shlee1@kaist.ac.kr
https://sites.google.com/site/leelab2013/
Sensory Processing Lab (SPL)
Department of Biological Sciences (BIO)
Korea Advanced Institute of Science and Technology (KAIST)
Profile:
You-Hyang Song
Researcher (Ph.D.)
dbgidtm17@kaist.ac.kr
SPL, KAIST BIO
Profile:
Yang-Sun Hwang
Researcher (M.S.)
hys940129@kaist.ac.kr
SPL, KAIST BIO
(END)
2020.04.23 View 14780
A Study Finds Neuropeptide Somatostatin Enhances Visual Processing
Researchers have confirmed that neuropeptide somatostatin can improve cognitive function in the brain. A research group of Professor Seung-Hee Lee from the Department of Biological Sciences at KAIST found that the application of neuropeptide somatostatin improves visual processing and cognitive behaviors by reducing excitatory inputs to parvalbumin-positive interneurons in the cortex.
This study, reported at Science Advances on April 22nd (EST), sheds a new light on the therapeutics of neurodegenerative diseases. According to a recent study in Korea, one in ten seniors over 65 is experiencing dementia-related symptoms in their daily lives such like memory loss, cognitive decline, and motion function disorders. Professor Lee believes that somatostatin treatment can be directly applied to the recovery of cognitive functions in Alzheimer’s disease patients.
Professor Lee started this study noting the fact that the level of somatostatin expression was dramatically decreased in the cerebral cortex and cerebrospinal fluid of Alzheimer’s disease patients
Somatostatin-expressing neurons in the cortex are known to exert the dendritic inhibition of pyramidal neurons via GABAergic transmission. Previous studies focused on their inhibitory effects on cortical circuits, but somatostatin-expressing neurons can co-release somatostatin upon activation. Despite the abundant expression of somatostatin and its receptors in the cerebral cortex, it was not known if somatostatin could modulate cognitive processing in the cortex.
The research team demonstrated that the somatostatin treatment into the cerebral cortex could enhance visual processing and cognitive behaviors in mice. The research team combined behaviors, in vivo and in vitro electrophysiology, and electron microscopy techniques to reveal how the activation of somatostatin receptors in vivo enhanced the ability of visual recognition in animals. Interestingly, somatostatin release can reduce excitatory synaptic transmission to another subtype of GABAergic interneurons, parvalbumin (PV)-expressing neurons.
As somatostatin is a stable and safe neuropeptide expressed naturally in the mammalian brain, it was safe to be injected into the cortex and cerebrospinal fluid, showing a potential application to drug development for curing cognitive disorders in humans.
Professor Lee said, “Our research confirmed the key role of the neuropeptide SST in modulating cortical function and enhancing cognitive ability in the mammalian brain. I hope new drugs can be developed based on the function of somatostatin to treat cognitive disabilities in many patients suffering from neurological disorders.”
This study was supported by the National Research Foundation of Korea.
Publication:
Song, Y. H et al. (2020) ‘Somatostatin enhances visual processing and perception by suppressing excitatory inputs to parvalbumin-positive interneurons in V1’, Science Advances, 6(17). Available online at https://doi.org/10.1126/sciadv.aaz0517
Profile:
Seung-Hee Lee
Associate Professor
shlee1@kaist.ac.kr
https://sites.google.com/site/leelab2013/
Sensory Processing Lab (SPL)
Department of Biological Sciences (BIO)
Korea Advanced Institute of Science and Technology (KAIST)
Profile:
You-Hyang Song
Researcher (Ph.D.)
dbgidtm17@kaist.ac.kr
SPL, KAIST BIO
Profile:
Yang-Sun Hwang
Researcher (M.S.)
hys940129@kaist.ac.kr
SPL, KAIST BIO
(END)
2020.04.23 View 14780 -
 Cyber MOU Signing with Zhejiang University
KAIST signed an MOU with Zhejiang University (ZJU) in China on March 25. This MOU signing ceremony took place via video conference due to the outbreak of COVID-19.
The collaboration with ZJU had already started with the signing of an MOU for cooperation in technology commercialization last December. Possible cooperation initiatives included facilitating joint start-up businesses, patent portfolios, and technology marketing.
With this general agreement signing, it is expected that the two institutes will expand mutual exchanges and collaborations at the institutional level for education and research.
President Sung-Chul Shin said, “We will work together to devise measures for the systematic advancement of cooperation in various directions, including education, research, and the commercialization of technologies.”
ZJU, a member of the C9 League known as China’s Ivy League, was established in 1897 and is located in the city of Hangzhou. Its population across 37 colleges and schools comprises 54,641 students and 3,741 faculty members. The university was ranked 6th in Asia and 54th in the world in the 2020 QS Rankings.
(END)
2020.03.30 View 15178
Cyber MOU Signing with Zhejiang University
KAIST signed an MOU with Zhejiang University (ZJU) in China on March 25. This MOU signing ceremony took place via video conference due to the outbreak of COVID-19.
The collaboration with ZJU had already started with the signing of an MOU for cooperation in technology commercialization last December. Possible cooperation initiatives included facilitating joint start-up businesses, patent portfolios, and technology marketing.
With this general agreement signing, it is expected that the two institutes will expand mutual exchanges and collaborations at the institutional level for education and research.
President Sung-Chul Shin said, “We will work together to devise measures for the systematic advancement of cooperation in various directions, including education, research, and the commercialization of technologies.”
ZJU, a member of the C9 League known as China’s Ivy League, was established in 1897 and is located in the city of Hangzhou. Its population across 37 colleges and schools comprises 54,641 students and 3,741 faculty members. The university was ranked 6th in Asia and 54th in the world in the 2020 QS Rankings.
(END)
2020.03.30 View 15178 -
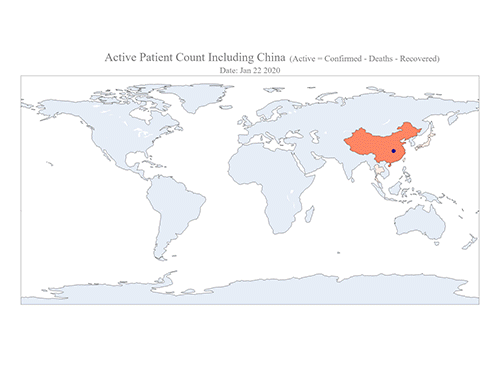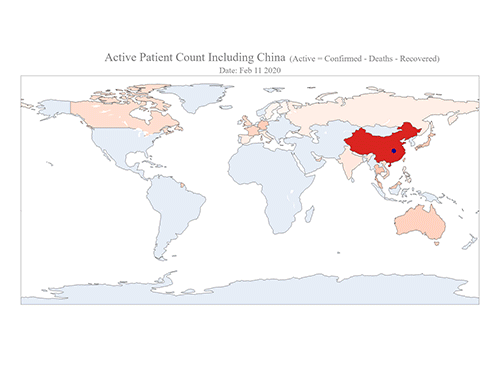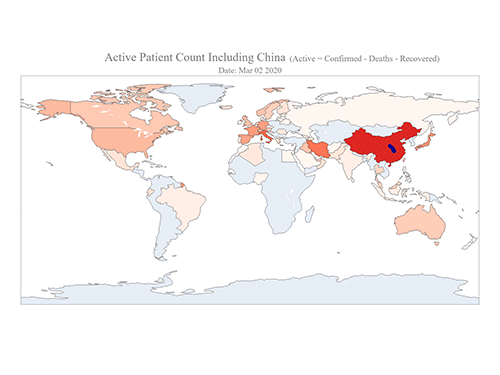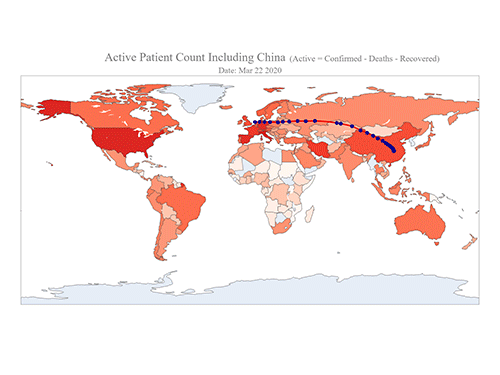 COVID-19 Map Shows How the Global Pandemic Moves
- A School of Computing team facilitated the data from COVID-19 to show the global spread of the virus. -
The COVID-19 map made by KAIST data scientists shows where and how the virus is spreading from China, reportedly the epicenter of the disease.
Professor Meeyoung Cha from the School of Computing and her group facilitated data based on the number of confirmed cases from January 22 to March 22 to analyze the trends of this global epidemic. The statistics include the number of confirmed cases, recoveries, and deaths across major continents based on the number of confirmed case data during that period.
The moving dot on the map strikingly shows how the confirmed cases are moving across the globe. According to their statistics, the centroid of the disease starts from near Wuhan in China and moved to Korea, then through the European region via Italy and Iran.
The data is collected by a graduate student from the School of Computing, Geng Sun, who started the process during the time he was quarantined since coming back from his home in China. An undergraduate colleague of Geng's, Gabriel Camilo Lima who made the map, is now working remotely from his home in Brazil since all undergraduate students were required to move out of the dormitory last week. The university closed all undergraduate housing and advised the undergraduate students to go back home in a preventive measure to stop the virus from spreading across the campus.
Gabriel said he calculated the centroid of all confirmed cases up to a given day. He explained, “I weighed each coordinate by the number of cases in that region and country and calculated an approximate center of gravity.”
“The Earth is round, so the shortest path from Asia to Europe is often through Russia. In early March, the center of gravity of new cases was moving from Asia to Europe. Therefore, the centroid is moving to the west and goes through Russia, even though Russia has not reported many cases,” he added.
Professor Cha, who is also responsible for the Data Science Group at the Institute for Basic Science (IBS) as the Chief Investigator, said their group will continue to update the map using public data at https://ds.ibs.re.kr/index.php/covid-19/.
(END)
2020.03.27 View 13882
COVID-19 Map Shows How the Global Pandemic Moves
- A School of Computing team facilitated the data from COVID-19 to show the global spread of the virus. -
The COVID-19 map made by KAIST data scientists shows where and how the virus is spreading from China, reportedly the epicenter of the disease.
Professor Meeyoung Cha from the School of Computing and her group facilitated data based on the number of confirmed cases from January 22 to March 22 to analyze the trends of this global epidemic. The statistics include the number of confirmed cases, recoveries, and deaths across major continents based on the number of confirmed case data during that period.
The moving dot on the map strikingly shows how the confirmed cases are moving across the globe. According to their statistics, the centroid of the disease starts from near Wuhan in China and moved to Korea, then through the European region via Italy and Iran.
The data is collected by a graduate student from the School of Computing, Geng Sun, who started the process during the time he was quarantined since coming back from his home in China. An undergraduate colleague of Geng's, Gabriel Camilo Lima who made the map, is now working remotely from his home in Brazil since all undergraduate students were required to move out of the dormitory last week. The university closed all undergraduate housing and advised the undergraduate students to go back home in a preventive measure to stop the virus from spreading across the campus.
Gabriel said he calculated the centroid of all confirmed cases up to a given day. He explained, “I weighed each coordinate by the number of cases in that region and country and calculated an approximate center of gravity.”
“The Earth is round, so the shortest path from Asia to Europe is often through Russia. In early March, the center of gravity of new cases was moving from Asia to Europe. Therefore, the centroid is moving to the west and goes through Russia, even though Russia has not reported many cases,” he added.
Professor Cha, who is also responsible for the Data Science Group at the Institute for Basic Science (IBS) as the Chief Investigator, said their group will continue to update the map using public data at https://ds.ibs.re.kr/index.php/covid-19/.
(END)
2020.03.27 View 13882