computational
-
 KAIST Proposes a New Way to Circumvent a Long-time Frustration in Neural Computing
The human brain begins learning through spontaneous random activities even before it receives sensory information from the external world. The technology developed by the KAIST research team enables much faster and more accurate learning when exposed to actual data by pre-learning random information in a brain-mimicking artificial neural network, and is expected to be a breakthrough in the development of brain-based artificial intelligence and neuromorphic computing technology in the future.
KAIST (President Kwang-Hyung Lee) announced on the 16th of December that Professor Se-Bum Paik 's research team in the Department of Brain Cognitive Sciences solved the weight transport problem*, a long-standing challenge in neural network learning, and through this, explained the principles that enable resource-efficient learning in biological brain neural networks.
*Weight transport problem: This is the biggest obstacle to the development of artificial intelligence that mimics the biological brain. It is the fundamental reason why large-scale memory and computational work are required in the learning of general artificial neural networks, unlike biological brains.
Over the past several decades, the development of artificial intelligence has been based on error backpropagation learning proposed by Geoffery Hinton, who won the Nobel Prize in Physics this year. However, error backpropagation learning was thought to be impossible in biological brains because it requires the unrealistic assumption that individual neurons must know all the connected information across multiple layers in order to calculate the error signal for learning.
< Figure 1. Illustration depicting the method of random noise training and its effects >
This difficult problem, called the weight transport problem, was raised by Francis Crick, who won the Nobel Prize in Physiology or Medicine for the discovery of the structure of DNA, after the error backpropagation learning was proposed by Hinton in 1986. Since then, it has been considered the reason why the operating principles of natural neural networks and artificial neural networks will forever be fundamentally different.
At the borderline of artificial intelligence and neuroscience, researchers including Hinton have continued to attempt to create biologically plausible models that can implement the learning principles of the brain by solving the weight transport problem.
In 2016, a joint research team from Oxford University and DeepMind in the UK first proposed the concept of error backpropagation learning being possible without weight transport, drawing attention from the academic world. However, biologically plausible error backpropagation learning without weight transport was inefficient, with slow learning speeds and low accuracy, making it difficult to apply in reality.
KAIST research team noted that the biological brain begins learning through internal spontaneous random neural activity even before experiencing external sensory experiences. To mimic this, the research team pre-trained a biologically plausible neural network without weight transport with meaningless random information (random noise).
As a result, they showed that the symmetry of the forward and backward neural cell connections of the neural network, which is an essential condition for error backpropagation learning, can be created. In other words, learning without weight transport is possible through random pre-training.
< Figure 2. Illustration depicting the meta-learning effect of random noise training >
The research team revealed that learning random information before learning actual data has the property of meta-learning, which is ‘learning how to learn.’ It was shown that neural networks that pre-learned random noise perform much faster and more accurate learning when exposed to actual data, and can achieve high learning efficiency without weight transport.
< Figure 3. Illustration depicting research on understanding the brain's operating principles through artificial neural networks >
Professor Se-Bum Paik said, “It breaks the conventional understanding of existing machine learning that only data learning is important, and provides a new perspective that focuses on the neuroscience principles of creating appropriate conditions before learning,” and added, “It is significant in that it solves important problems in artificial neural network learning through clues from developmental neuroscience, and at the same time provides insight into the brain’s learning principles through artificial neural network models.”
This study, in which Jeonghwan Cheon, a Master’s candidate of KAIST Department of Brain and Cognitive Sciences participated as the first author and Professor Sang Wan Lee of the same department as a co-author, was presented at the 38th Neural Information Processing Systems (NeurIPS), the world's top artificial intelligence conference, on December 14th in Vancouver, Canada. (Paper title: Pretraining with random noise for fast and robust learning without weight transport)
This study was conducted with the support of the National Research Foundation of Korea's Basic Research Program in Science and Engineering, the Information and Communications Technology Planning and Evaluation Institute's Talent Development Program, and the KAIST Singularity Professor Program.
2024.12.16 View 7031
KAIST Proposes a New Way to Circumvent a Long-time Frustration in Neural Computing
The human brain begins learning through spontaneous random activities even before it receives sensory information from the external world. The technology developed by the KAIST research team enables much faster and more accurate learning when exposed to actual data by pre-learning random information in a brain-mimicking artificial neural network, and is expected to be a breakthrough in the development of brain-based artificial intelligence and neuromorphic computing technology in the future.
KAIST (President Kwang-Hyung Lee) announced on the 16th of December that Professor Se-Bum Paik 's research team in the Department of Brain Cognitive Sciences solved the weight transport problem*, a long-standing challenge in neural network learning, and through this, explained the principles that enable resource-efficient learning in biological brain neural networks.
*Weight transport problem: This is the biggest obstacle to the development of artificial intelligence that mimics the biological brain. It is the fundamental reason why large-scale memory and computational work are required in the learning of general artificial neural networks, unlike biological brains.
Over the past several decades, the development of artificial intelligence has been based on error backpropagation learning proposed by Geoffery Hinton, who won the Nobel Prize in Physics this year. However, error backpropagation learning was thought to be impossible in biological brains because it requires the unrealistic assumption that individual neurons must know all the connected information across multiple layers in order to calculate the error signal for learning.
< Figure 1. Illustration depicting the method of random noise training and its effects >
This difficult problem, called the weight transport problem, was raised by Francis Crick, who won the Nobel Prize in Physiology or Medicine for the discovery of the structure of DNA, after the error backpropagation learning was proposed by Hinton in 1986. Since then, it has been considered the reason why the operating principles of natural neural networks and artificial neural networks will forever be fundamentally different.
At the borderline of artificial intelligence and neuroscience, researchers including Hinton have continued to attempt to create biologically plausible models that can implement the learning principles of the brain by solving the weight transport problem.
In 2016, a joint research team from Oxford University and DeepMind in the UK first proposed the concept of error backpropagation learning being possible without weight transport, drawing attention from the academic world. However, biologically plausible error backpropagation learning without weight transport was inefficient, with slow learning speeds and low accuracy, making it difficult to apply in reality.
KAIST research team noted that the biological brain begins learning through internal spontaneous random neural activity even before experiencing external sensory experiences. To mimic this, the research team pre-trained a biologically plausible neural network without weight transport with meaningless random information (random noise).
As a result, they showed that the symmetry of the forward and backward neural cell connections of the neural network, which is an essential condition for error backpropagation learning, can be created. In other words, learning without weight transport is possible through random pre-training.
< Figure 2. Illustration depicting the meta-learning effect of random noise training >
The research team revealed that learning random information before learning actual data has the property of meta-learning, which is ‘learning how to learn.’ It was shown that neural networks that pre-learned random noise perform much faster and more accurate learning when exposed to actual data, and can achieve high learning efficiency without weight transport.
< Figure 3. Illustration depicting research on understanding the brain's operating principles through artificial neural networks >
Professor Se-Bum Paik said, “It breaks the conventional understanding of existing machine learning that only data learning is important, and provides a new perspective that focuses on the neuroscience principles of creating appropriate conditions before learning,” and added, “It is significant in that it solves important problems in artificial neural network learning through clues from developmental neuroscience, and at the same time provides insight into the brain’s learning principles through artificial neural network models.”
This study, in which Jeonghwan Cheon, a Master’s candidate of KAIST Department of Brain and Cognitive Sciences participated as the first author and Professor Sang Wan Lee of the same department as a co-author, was presented at the 38th Neural Information Processing Systems (NeurIPS), the world's top artificial intelligence conference, on December 14th in Vancouver, Canada. (Paper title: Pretraining with random noise for fast and robust learning without weight transport)
This study was conducted with the support of the National Research Foundation of Korea's Basic Research Program in Science and Engineering, the Information and Communications Technology Planning and Evaluation Institute's Talent Development Program, and the KAIST Singularity Professor Program.
2024.12.16 View 7031 -
 KAIST Employs Image-recognition AI to Determine Battery Composition and Conditions
An international collaborative research team has developed an image recognition technology that can accurately determine the elemental composition and the number of charge and discharge cycles of a battery by examining only its surface morphology using AI learning.
KAIST (President Kwang-Hyung Lee) announced on July 2nd that Professor Seungbum Hong from the Department of Materials Science and Engineering, in collaboration with the Electronics and Telecommunications Research Institute (ETRI) and Drexel University in the United States, has developed a method to predict the major elemental composition and charge-discharge state of NCM cathode materials with 99.6% accuracy using convolutional neural networks (CNN)*.
*Convolutional Neural Network (CNN): A type of multi-layer, feed-forward, artificial neural network used for analyzing visual images.
The research team noted that while scanning electron microscopy (SEM) is used in semiconductor manufacturing to inspect wafer defects, it is rarely used in battery inspections. SEM is used for batteries to analyze the size of particles only at research sites, and reliability is predicted from the broken particles and the shape of the breakage in the case of deteriorated battery materials.
The research team decided that it would be groundbreaking if an automated SEM can be used in the process of battery production, just like in the semiconductor manufacturing, to inspect the surface of the cathode material to determine whether it was synthesized according to the desired composition and that the lifespan would be reliable, thereby reducing the defect rate.
< Figure 1. Example images of true cases and their grad-CAM overlays from the best trained network. >
The researchers trained a CNN-based AI applicable to autonomous vehicles to learn the surface images of battery materials, enabling it to predict the major elemental composition and charge-discharge cycle states of the cathode materials. They found that while the method could accurately predict the composition of materials with additives, it had lower accuracy for predicting charge-discharge states. The team plans to further train the AI with various battery material morphologies produced through different processes and ultimately use it for inspecting the compositional uniformity and predicting the lifespan of next-generation batteries.
Professor Joshua C. Agar, one of the collaborating researchers of the project from the Department of Mechanical Engineering and Mechanics of Drexel University, said, "In the future, artificial intelligence is expected to be applied not only to battery materials but also to various dynamic processes in functional materials synthesis, clean energy generation in fusion, and understanding foundations of particles and the universe."
Professor Seungbum Hong from KAIST, who led the research, stated, "This research is significant as it is the first in the world to develop an AI-based methodology that can quickly and accurately predict the major elemental composition and the state of the battery from the structural data of micron-scale SEM images. The methodology developed in this study for identifying the composition and state of battery materials based on microscopic images is expected to play a crucial role in improving the performance and quality of battery materials in the future."
< Figure 2. Accuracies of CNN Model predictions on SEM images of NCM cathode materials with additives under various conditions. >
This research was conducted by KAIST’s Materials Science and Engineering Department graduates Dr. Jimin Oh and Dr. Jiwon Yeom, the co-first authors, in collaboration with Professor Josh Agar and Dr. Kwang Man Kim from ETRI. It was supported by the National Research Foundation of Korea, the KAIST Global Singularity project, and international collaboration with the US research team. The results were published in the international journal npj Computational Materials on May 4. (Paper Title: “Composition and state prediction of lithium-ion cathode via convolutional neural network trained on scanning electron microscopy images”)
2024.07.02 View 6588
KAIST Employs Image-recognition AI to Determine Battery Composition and Conditions
An international collaborative research team has developed an image recognition technology that can accurately determine the elemental composition and the number of charge and discharge cycles of a battery by examining only its surface morphology using AI learning.
KAIST (President Kwang-Hyung Lee) announced on July 2nd that Professor Seungbum Hong from the Department of Materials Science and Engineering, in collaboration with the Electronics and Telecommunications Research Institute (ETRI) and Drexel University in the United States, has developed a method to predict the major elemental composition and charge-discharge state of NCM cathode materials with 99.6% accuracy using convolutional neural networks (CNN)*.
*Convolutional Neural Network (CNN): A type of multi-layer, feed-forward, artificial neural network used for analyzing visual images.
The research team noted that while scanning electron microscopy (SEM) is used in semiconductor manufacturing to inspect wafer defects, it is rarely used in battery inspections. SEM is used for batteries to analyze the size of particles only at research sites, and reliability is predicted from the broken particles and the shape of the breakage in the case of deteriorated battery materials.
The research team decided that it would be groundbreaking if an automated SEM can be used in the process of battery production, just like in the semiconductor manufacturing, to inspect the surface of the cathode material to determine whether it was synthesized according to the desired composition and that the lifespan would be reliable, thereby reducing the defect rate.
< Figure 1. Example images of true cases and their grad-CAM overlays from the best trained network. >
The researchers trained a CNN-based AI applicable to autonomous vehicles to learn the surface images of battery materials, enabling it to predict the major elemental composition and charge-discharge cycle states of the cathode materials. They found that while the method could accurately predict the composition of materials with additives, it had lower accuracy for predicting charge-discharge states. The team plans to further train the AI with various battery material morphologies produced through different processes and ultimately use it for inspecting the compositional uniformity and predicting the lifespan of next-generation batteries.
Professor Joshua C. Agar, one of the collaborating researchers of the project from the Department of Mechanical Engineering and Mechanics of Drexel University, said, "In the future, artificial intelligence is expected to be applied not only to battery materials but also to various dynamic processes in functional materials synthesis, clean energy generation in fusion, and understanding foundations of particles and the universe."
Professor Seungbum Hong from KAIST, who led the research, stated, "This research is significant as it is the first in the world to develop an AI-based methodology that can quickly and accurately predict the major elemental composition and the state of the battery from the structural data of micron-scale SEM images. The methodology developed in this study for identifying the composition and state of battery materials based on microscopic images is expected to play a crucial role in improving the performance and quality of battery materials in the future."
< Figure 2. Accuracies of CNN Model predictions on SEM images of NCM cathode materials with additives under various conditions. >
This research was conducted by KAIST’s Materials Science and Engineering Department graduates Dr. Jimin Oh and Dr. Jiwon Yeom, the co-first authors, in collaboration with Professor Josh Agar and Dr. Kwang Man Kim from ETRI. It was supported by the National Research Foundation of Korea, the KAIST Global Singularity project, and international collaboration with the US research team. The results were published in the international journal npj Computational Materials on May 4. (Paper Title: “Composition and state prediction of lithium-ion cathode via convolutional neural network trained on scanning electron microscopy images”)
2024.07.02 View 6588 -
 KAIST leads AI-based analysis on drug-drug interactions involving Paxlovid
KAIST (President Kwang Hyung Lee) announced on the 16th that an advanced AI-based drug interaction prediction technology developed by the Distinguished Professor Sang Yup Lee's research team in the Department of Biochemical Engineering that analyzed the interaction between the PaxlovidTM ingredients that are used as COVID-19 treatment and other prescription drugs was published as a thesis. This paper was published in the online edition of 「Proceedings of the National Academy of Sciences of America」 (PNAS), an internationally renowned academic journal, on the 13th of March.
* Thesis Title: Computational prediction of interactions between Paxlovid and prescription drugs (Authored by Yeji Kim (KAIST, co-first author), Jae Yong Ryu (Duksung Women's University, co-first author), Hyun Uk Kim (KAIST, co-first author), and Sang Yup Lee (KAIST, corresponding author))
In this study, the research team developed DeepDDI2, an advanced version of DeepDDI, an AI-based drug interaction prediction model they developed in 2018. DeepDDI2 is able to compute for and process a total of 113 drug-drug interaction (DDI) types, more than the 86 DDI types covered by the existing DeepDDI.
The research team used DeepDDI2 to predict possible interactions between the ingredients (ritonavir, nirmatrelvir) of Paxlovid*, a COVID-19 treatment, and other prescription drugs. The research team said that while among COVID-19 patients, high-risk patients with chronic diseases such as high blood pressure and diabetes are likely to be taking other drugs, drug-drug interactions and adverse drug reactions for Paxlovid have not been sufficiently analyzed, yet. This study was pursued in light of seeing how continued usage of the drug may lead to serious and unwanted complications.
* Paxlovid: Paxlovid is a COVID-19 treatment developed by Pfizer, an American pharmaceutical company, and received emergency use approval (EUA) from the US Food and Drug Administration (FDA) in December 2021.
The research team used DeepDDI2 to predict how Paxrovid's components, ritonavir and nirmatrelvir, would interact with 2,248 prescription drugs. As a result of the prediction, ritonavir was predicted to interact with 1,403 prescription drugs and nirmatrelvir with 673 drugs.
Using the prediction results, the research team proposed alternative drugs with the same mechanism but low drug interaction potential for prescription drugs with high adverse drug events (ADEs). Accordingly, 124 alternative drugs that could reduce the possible adverse DDI with ritonavir and 239 alternative drugs for nirmatrelvir were identified.
Through this research achievement, it became possible to use an deep learning technology to accurately predict drug-drug interactions (DDIs), and this is expected to play an important role in the digital healthcare, precision medicine and pharmaceutical industries by providing useful information in the process of developing new drugs and making prescriptions.
Distinguished Professor Sang Yup Lee said, "The results of this study are meaningful at times like when we would have to resort to using drugs that are developed in a hurry in the face of an urgent situations like the COVID-19 pandemic, that it is now possible to identify and take necessary actions against adverse drug reactions caused by drug-drug interactions very quickly.”
This research was carried out with the support of the KAIST New-Deal Project for COVID-19 Science and Technology and the Bio·Medical Technology Development Project supported by the Ministry of Science and ICT.
Figure 1. Results of drug interaction prediction between Paxlovid ingredients and representative approved drugs using DeepDDI2
2023.03.16 View 9398
KAIST leads AI-based analysis on drug-drug interactions involving Paxlovid
KAIST (President Kwang Hyung Lee) announced on the 16th that an advanced AI-based drug interaction prediction technology developed by the Distinguished Professor Sang Yup Lee's research team in the Department of Biochemical Engineering that analyzed the interaction between the PaxlovidTM ingredients that are used as COVID-19 treatment and other prescription drugs was published as a thesis. This paper was published in the online edition of 「Proceedings of the National Academy of Sciences of America」 (PNAS), an internationally renowned academic journal, on the 13th of March.
* Thesis Title: Computational prediction of interactions between Paxlovid and prescription drugs (Authored by Yeji Kim (KAIST, co-first author), Jae Yong Ryu (Duksung Women's University, co-first author), Hyun Uk Kim (KAIST, co-first author), and Sang Yup Lee (KAIST, corresponding author))
In this study, the research team developed DeepDDI2, an advanced version of DeepDDI, an AI-based drug interaction prediction model they developed in 2018. DeepDDI2 is able to compute for and process a total of 113 drug-drug interaction (DDI) types, more than the 86 DDI types covered by the existing DeepDDI.
The research team used DeepDDI2 to predict possible interactions between the ingredients (ritonavir, nirmatrelvir) of Paxlovid*, a COVID-19 treatment, and other prescription drugs. The research team said that while among COVID-19 patients, high-risk patients with chronic diseases such as high blood pressure and diabetes are likely to be taking other drugs, drug-drug interactions and adverse drug reactions for Paxlovid have not been sufficiently analyzed, yet. This study was pursued in light of seeing how continued usage of the drug may lead to serious and unwanted complications.
* Paxlovid: Paxlovid is a COVID-19 treatment developed by Pfizer, an American pharmaceutical company, and received emergency use approval (EUA) from the US Food and Drug Administration (FDA) in December 2021.
The research team used DeepDDI2 to predict how Paxrovid's components, ritonavir and nirmatrelvir, would interact with 2,248 prescription drugs. As a result of the prediction, ritonavir was predicted to interact with 1,403 prescription drugs and nirmatrelvir with 673 drugs.
Using the prediction results, the research team proposed alternative drugs with the same mechanism but low drug interaction potential for prescription drugs with high adverse drug events (ADEs). Accordingly, 124 alternative drugs that could reduce the possible adverse DDI with ritonavir and 239 alternative drugs for nirmatrelvir were identified.
Through this research achievement, it became possible to use an deep learning technology to accurately predict drug-drug interactions (DDIs), and this is expected to play an important role in the digital healthcare, precision medicine and pharmaceutical industries by providing useful information in the process of developing new drugs and making prescriptions.
Distinguished Professor Sang Yup Lee said, "The results of this study are meaningful at times like when we would have to resort to using drugs that are developed in a hurry in the face of an urgent situations like the COVID-19 pandemic, that it is now possible to identify and take necessary actions against adverse drug reactions caused by drug-drug interactions very quickly.”
This research was carried out with the support of the KAIST New-Deal Project for COVID-19 Science and Technology and the Bio·Medical Technology Development Project supported by the Ministry of Science and ICT.
Figure 1. Results of drug interaction prediction between Paxlovid ingredients and representative approved drugs using DeepDDI2
2023.03.16 View 9398 -
 PICASSO Technique Drives Biological Molecules into Technicolor
The new imaging approach brings current imaging colors from four to more than 15 for mapping overlapping proteins
Pablo Picasso’s surreal cubist artistic style shifted common features into unrecognizable scenes, but a new imaging approach bearing his namesake may elucidate the most complicated subject: the brain. Employing artificial intelligence to clarify spectral color blending of tiny molecules used to stain specific proteins and other items of research interest, the PICASSO technique, allows researchers to use more than 15 colors to image and parse our overlapping proteins.
The PICASSO developers, based in Korea, published their approach on May 5 in Nature Communications.
Fluorophores — the staining molecules — emit specific colors when excited by a light, but if more than four fluorophores are used, their emitted colors overlap and blend. Researchers previously developed techniques to correct this spectral overlap by precisely defining the matrix of mixed and unmixed images. This measurement depends on reference spectra, found by identifying clear images of only one fluorophore-stained specimen or of multiple, identically prepared specimens that only contain a single fluorophore each.
“Such reference spectra measurement could be complicated to perform in highly heterogeneous specimens, such as the brain, due to the highly varied emission spectra of fluorophores depending on the subregions from which the spectra were measured,” said co-corresponding author Young-Gyu Yoon, professor in the School of Electrical Engineering at KAIST. He explained that the subregions would each need their own spectra reference measurements, making for an inefficient, time-consuming process. “To address this problem, we developed an approach that does not require reference spectra measurements.”
The approach is the “Process of ultra-multiplexed Imaging of biomolecules viA the unmixing of the Signals of Spectrally Overlapping fluorophores,” also known as PICASSO. Ultra-multiplexed imaging refers to visualizing the numerous individual components of a unit. Like a cinema multiplex in which each theater plays a different movie, each protein in a cell has a different role. By staining with fluorophores, researchers can begin to understand those roles.
“We devised a strategy based on information theory; unmixing is performed by iteratively minimizing the mutual information between mixed images,” said co-corresponding author Jae-Byum Chang, professor in the Department of Materials Science and Engineering, KAIST. “This allows us to get away with the assumption that the spatial distribution of different proteins is mutually exclusive and enables accurate information unmixing.”
To demonstrate PICASSO’s capabilities, the researchers applied the technique to imaging a mouse brain. With a single round of staining, they performed 15-color multiplexed imaging of a mouse brain. Although small, mouse brains are still complex, multifaceted organs that can take significant resources to map. According to the researchers, PICASSO can improve the capabilities of other imaging techniques and allow for the use of even more fluorophore colors.
Using one such imaging technique in combination with PICASSO, the team achieved 45-color multiplexed imaging of the mouse brain in only three staining and imaging cycles, according to Yoon.
“PICASSO is a versatile tool for the multiplexed biomolecule imaging of cultured cells, tissue slices and clinical specimens,” Chang said. “We anticipate that PICASSO will be useful for a broad range of applications for which biomolecules’ spatial information is important. One such application the tool would be useful for is revealing the cellular heterogeneities of tumor microenvironments, especially the heterogeneous populations of immune cells, which are closely related to cancer prognoses and the efficacy of cancer therapies.”
The Samsung Research Funding & Incubation Center for Future Technology supported this work. Spectral imaging was performed at the Korea Basic Science Institute Western Seoul Center.
-PublicationJunyoung Seo, Yeonbo Sim, Jeewon Kim, Hyunwoo Kim, In Cho, Hoyeon Nam, Yong-Gyu Yoon, Jae-Byum Chang, “PICASSO allows ultra-multiplexed fluorescence imaging of spatiallyoverlapping proteins without reference spectra measurements,” May 5, Nature Communications (doi.org/10.1038/s41467-022-30168-z)
-ProfileProfessor Jae-Byum ChangDepartment of Materials Science and EngineeringCollege of EngineeringKAIST
Professor Young-Gyu YoonSchool of Electrical EngineeringCollege of EngineeringKAIST
2022.06.22 View 11264
PICASSO Technique Drives Biological Molecules into Technicolor
The new imaging approach brings current imaging colors from four to more than 15 for mapping overlapping proteins
Pablo Picasso’s surreal cubist artistic style shifted common features into unrecognizable scenes, but a new imaging approach bearing his namesake may elucidate the most complicated subject: the brain. Employing artificial intelligence to clarify spectral color blending of tiny molecules used to stain specific proteins and other items of research interest, the PICASSO technique, allows researchers to use more than 15 colors to image and parse our overlapping proteins.
The PICASSO developers, based in Korea, published their approach on May 5 in Nature Communications.
Fluorophores — the staining molecules — emit specific colors when excited by a light, but if more than four fluorophores are used, their emitted colors overlap and blend. Researchers previously developed techniques to correct this spectral overlap by precisely defining the matrix of mixed and unmixed images. This measurement depends on reference spectra, found by identifying clear images of only one fluorophore-stained specimen or of multiple, identically prepared specimens that only contain a single fluorophore each.
“Such reference spectra measurement could be complicated to perform in highly heterogeneous specimens, such as the brain, due to the highly varied emission spectra of fluorophores depending on the subregions from which the spectra were measured,” said co-corresponding author Young-Gyu Yoon, professor in the School of Electrical Engineering at KAIST. He explained that the subregions would each need their own spectra reference measurements, making for an inefficient, time-consuming process. “To address this problem, we developed an approach that does not require reference spectra measurements.”
The approach is the “Process of ultra-multiplexed Imaging of biomolecules viA the unmixing of the Signals of Spectrally Overlapping fluorophores,” also known as PICASSO. Ultra-multiplexed imaging refers to visualizing the numerous individual components of a unit. Like a cinema multiplex in which each theater plays a different movie, each protein in a cell has a different role. By staining with fluorophores, researchers can begin to understand those roles.
“We devised a strategy based on information theory; unmixing is performed by iteratively minimizing the mutual information between mixed images,” said co-corresponding author Jae-Byum Chang, professor in the Department of Materials Science and Engineering, KAIST. “This allows us to get away with the assumption that the spatial distribution of different proteins is mutually exclusive and enables accurate information unmixing.”
To demonstrate PICASSO’s capabilities, the researchers applied the technique to imaging a mouse brain. With a single round of staining, they performed 15-color multiplexed imaging of a mouse brain. Although small, mouse brains are still complex, multifaceted organs that can take significant resources to map. According to the researchers, PICASSO can improve the capabilities of other imaging techniques and allow for the use of even more fluorophore colors.
Using one such imaging technique in combination with PICASSO, the team achieved 45-color multiplexed imaging of the mouse brain in only three staining and imaging cycles, according to Yoon.
“PICASSO is a versatile tool for the multiplexed biomolecule imaging of cultured cells, tissue slices and clinical specimens,” Chang said. “We anticipate that PICASSO will be useful for a broad range of applications for which biomolecules’ spatial information is important. One such application the tool would be useful for is revealing the cellular heterogeneities of tumor microenvironments, especially the heterogeneous populations of immune cells, which are closely related to cancer prognoses and the efficacy of cancer therapies.”
The Samsung Research Funding & Incubation Center for Future Technology supported this work. Spectral imaging was performed at the Korea Basic Science Institute Western Seoul Center.
-PublicationJunyoung Seo, Yeonbo Sim, Jeewon Kim, Hyunwoo Kim, In Cho, Hoyeon Nam, Yong-Gyu Yoon, Jae-Byum Chang, “PICASSO allows ultra-multiplexed fluorescence imaging of spatiallyoverlapping proteins without reference spectra measurements,” May 5, Nature Communications (doi.org/10.1038/s41467-022-30168-z)
-ProfileProfessor Jae-Byum ChangDepartment of Materials Science and EngineeringCollege of EngineeringKAIST
Professor Young-Gyu YoonSchool of Electrical EngineeringCollege of EngineeringKAIST
2022.06.22 View 11264 -
 Scientist Discover How Circadian Rhythm Can Be Both Strong and Flexible
Study reveals that master and slave oscillators function via different molecular mechanisms
From tiny fruit flies to human beings, all animals on Earth maintain their daily rhythms based on their internal circadian clock. The circadian clock enables organisms to undergo rhythmic changes in behavior and physiology based on a 24-hour circadian cycle. For example, our own biological clock tells our brain to release melatonin, a sleep-inducing hormone, at night time.
The discovery of the molecular mechanism of the circadian clock was bestowed the Nobel Prize in Physiology or Medicine 2017. From what we know, no one centralized clock is responsible for our circadian cycles. Instead, it operates in a hierarchical network where there are “master pacemaker” and “slave oscillator”.
The master pacemaker receives various input signals from the environment such as light. The master then drives the slave oscillator that regulates various outputs such as sleep, feeding, and metabolism. Despite the different roles of the pacemaker neurons, they are known to share common molecular mechanisms that are well conserved in all lifeforms. For example, interlocked systems of multiple transcriptional-translational feedback loops (TTFLs) composed of core clock proteins have been deeply studied in fruit flies.
However, there is still much that we need to learn about our own biological clock. The hierarchically-organized nature of master and slave clock neurons leads to a prevailing belief that they share an identical molecular clockwork. At the same time, the different roles they serve in regulating bodily rhythms also raise the question of whether they might function under different molecular clockworks.
Research team led by Professor Kim Jae Kyoung from the Department of Mathematical Sciences, a chief investigator at the Biomedical Mathematics Group at the Institute for Basic Science, used a combination of mathematical and experimental approaches using fruit flies to answer this question. The team found that the master clock and the slave clock operate via different molecular mechanisms.
In both master and slave neurons of fruit flies, a circadian rhythm-related protein called PER is produced and degraded at different rates depending on the time of the day. Previously, the team found that the master clock neuron (sLNvs) and the slave clock neuron (DN1ps) have different profiles of PER in wild-type and Clk-Δ mutant Drosophila. This hinted that there might be a potential difference in molecular clockworks between the master and slave clock neurons.
However, due to the complexity of the molecular clockwork, it was challenging to identify the source of such differences. Thus, the team developed a mathematical model describing the molecular clockworks of the master and slave clocks. Then, all possible molecular differences between the master and slave clock neurons were systematically investigated by using computer simulations. The model predicted that PER is more efficiently produced and then rapidly degraded in the master clock compared to the slave clock neurons. This prediction was then confirmed by the follow-up experiments using animal.
Then, why do the master clock neurons have such different molecular properties from the slave clock neurons? To answer this question, the research team again used the combination of mathematical model simulation and experiments. It was found that the faster rate of synthesis of PER in the master clock neurons allows them to generate synchronized rhythms with a high level of amplitude. Generation of such a strong rhythm with high amplitude is critical to delivering clear signals to slave clock neurons.
However, such strong rhythms would typically be unfavorable when it comes to adapting to environmental changes. These include natural causes such as different daylight hours across summer and winter seasons, up to more extreme artificial cases such as jet lag that occurs after international travel. Thanks to the distinct property of the master clock neurons, it is able to undergo phase dispersion when the standard light-dark cycle is disrupted, drastically reducing the level of PER. The master clock neurons can then easily adapt to the new diurnal cycle. Our master pacemaker’s plasticity explains how we can quickly adjust to the new time zones after international flights after just a brief period of jet lag.
It is hoped that the findings of this study can have future clinical implications when it comes to treating various disorders that affect our circadian rhythm. Professor Kim notes, “When the circadian clock loses its robustness and flexibility, the circadian rhythms sleep disorders can occur. As this study identifies the molecular mechanism that generates robustness and flexibility of the circadian clock, it can facilitate the identification of the cause of and treatment strategy for the circadian rhythm sleep disorders.” This work was supported by the Human Frontier Science Program.
-PublicationEui Min Jeong, Miri Kwon, Eunjoo Cho, Sang Hyuk Lee, Hyun Kim, Eun Young Kim, and Jae Kyoung Kim, “Systematic modeling-driven experiments identify distinct molecularclockworks underlying hierarchically organized pacemaker neurons,” February 22, 2022, Proceedings of the National Academy of Sciences of the United States of America
-ProfileProfessor Jae Kyoung KimDepartment of Mathematical SciencesKAIST
2022.02.23 View 11265
Scientist Discover How Circadian Rhythm Can Be Both Strong and Flexible
Study reveals that master and slave oscillators function via different molecular mechanisms
From tiny fruit flies to human beings, all animals on Earth maintain their daily rhythms based on their internal circadian clock. The circadian clock enables organisms to undergo rhythmic changes in behavior and physiology based on a 24-hour circadian cycle. For example, our own biological clock tells our brain to release melatonin, a sleep-inducing hormone, at night time.
The discovery of the molecular mechanism of the circadian clock was bestowed the Nobel Prize in Physiology or Medicine 2017. From what we know, no one centralized clock is responsible for our circadian cycles. Instead, it operates in a hierarchical network where there are “master pacemaker” and “slave oscillator”.
The master pacemaker receives various input signals from the environment such as light. The master then drives the slave oscillator that regulates various outputs such as sleep, feeding, and metabolism. Despite the different roles of the pacemaker neurons, they are known to share common molecular mechanisms that are well conserved in all lifeforms. For example, interlocked systems of multiple transcriptional-translational feedback loops (TTFLs) composed of core clock proteins have been deeply studied in fruit flies.
However, there is still much that we need to learn about our own biological clock. The hierarchically-organized nature of master and slave clock neurons leads to a prevailing belief that they share an identical molecular clockwork. At the same time, the different roles they serve in regulating bodily rhythms also raise the question of whether they might function under different molecular clockworks.
Research team led by Professor Kim Jae Kyoung from the Department of Mathematical Sciences, a chief investigator at the Biomedical Mathematics Group at the Institute for Basic Science, used a combination of mathematical and experimental approaches using fruit flies to answer this question. The team found that the master clock and the slave clock operate via different molecular mechanisms.
In both master and slave neurons of fruit flies, a circadian rhythm-related protein called PER is produced and degraded at different rates depending on the time of the day. Previously, the team found that the master clock neuron (sLNvs) and the slave clock neuron (DN1ps) have different profiles of PER in wild-type and Clk-Δ mutant Drosophila. This hinted that there might be a potential difference in molecular clockworks between the master and slave clock neurons.
However, due to the complexity of the molecular clockwork, it was challenging to identify the source of such differences. Thus, the team developed a mathematical model describing the molecular clockworks of the master and slave clocks. Then, all possible molecular differences between the master and slave clock neurons were systematically investigated by using computer simulations. The model predicted that PER is more efficiently produced and then rapidly degraded in the master clock compared to the slave clock neurons. This prediction was then confirmed by the follow-up experiments using animal.
Then, why do the master clock neurons have such different molecular properties from the slave clock neurons? To answer this question, the research team again used the combination of mathematical model simulation and experiments. It was found that the faster rate of synthesis of PER in the master clock neurons allows them to generate synchronized rhythms with a high level of amplitude. Generation of such a strong rhythm with high amplitude is critical to delivering clear signals to slave clock neurons.
However, such strong rhythms would typically be unfavorable when it comes to adapting to environmental changes. These include natural causes such as different daylight hours across summer and winter seasons, up to more extreme artificial cases such as jet lag that occurs after international travel. Thanks to the distinct property of the master clock neurons, it is able to undergo phase dispersion when the standard light-dark cycle is disrupted, drastically reducing the level of PER. The master clock neurons can then easily adapt to the new diurnal cycle. Our master pacemaker’s plasticity explains how we can quickly adjust to the new time zones after international flights after just a brief period of jet lag.
It is hoped that the findings of this study can have future clinical implications when it comes to treating various disorders that affect our circadian rhythm. Professor Kim notes, “When the circadian clock loses its robustness and flexibility, the circadian rhythms sleep disorders can occur. As this study identifies the molecular mechanism that generates robustness and flexibility of the circadian clock, it can facilitate the identification of the cause of and treatment strategy for the circadian rhythm sleep disorders.” This work was supported by the Human Frontier Science Program.
-PublicationEui Min Jeong, Miri Kwon, Eunjoo Cho, Sang Hyuk Lee, Hyun Kim, Eun Young Kim, and Jae Kyoung Kim, “Systematic modeling-driven experiments identify distinct molecularclockworks underlying hierarchically organized pacemaker neurons,” February 22, 2022, Proceedings of the National Academy of Sciences of the United States of America
-ProfileProfessor Jae Kyoung KimDepartment of Mathematical SciencesKAIST
2022.02.23 View 11265 -
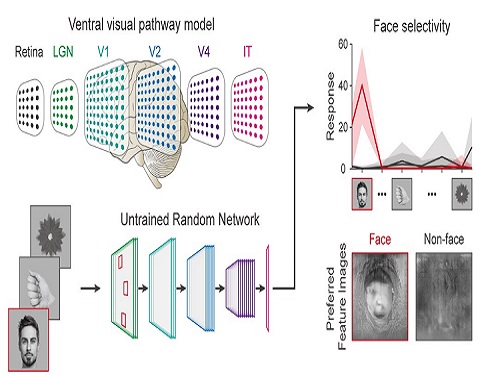 Face Detection in Untrained Deep Neural Networks
A KAIST team shows that primitive visual selectivity of faces can arise spontaneously in completely untrained deep neural networks
Researchers have found that higher visual cognitive functions can arise spontaneously in untrained neural networks. A KAIST research team led by Professor Se-Bum Paik from the Department of Bio and Brain Engineering has shown that visual selectivity of facial images can arise even in completely untrained deep neural networks.
This new finding has provided revelatory insights into mechanisms underlying the development of cognitive functions in both biological and artificial neural networks, also making a significant impact on our understanding of the origin of early brain functions before sensory experiences.
The study published in Nature Communications on December 16 demonstrates that neuronal activities selective to facial images are observed in randomly initialized deep neural networks in the complete absence of learning, and that they show the characteristics of those observed in biological brains.
The ability to identify and recognize faces is a crucial function for social behavior, and this ability is thought to originate from neuronal tuning at the single or multi-neuronal level. Neurons that selectively respond to faces are observed in young animals of various species, and this raises intense debate whether face-selective neurons can arise innately in the brain or if they require visual experience.
Using a model neural network that captures properties of the ventral stream of the visual cortex, the research team found that face-selectivity can emerge spontaneously from random feedforward wirings in untrained deep neural networks. The team showed that the character of this innate face-selectivity is comparable to that observed with face-selective neurons in the brain, and that this spontaneous neuronal tuning for faces enables the network to perform face detection tasks.
These results imply a possible scenario in which the random feedforward connections that develop in early, untrained networks may be sufficient for initializing primitive visual cognitive functions.
Professor Paik said, “Our findings suggest that innate cognitive functions can emerge spontaneously from the statistical complexity embedded in the hierarchical feedforward projection circuitry, even in the complete absence of learning”.
He continued, “Our results provide a broad conceptual advance as well as advanced insight into the mechanisms underlying the development of innate functions in both biological and artificial neural networks, which may unravel the mystery of the generation and evolution of intelligence.” This work was supported by the National Research Foundation of Korea (NRF) and by the KAIST singularity research project.
-PublicationSeungdae Baek, Min Song, Jaeson Jang, Gwangsu Kim, and Se-Bum Baik, “Face detection in untrained deep neural network,” Nature Communications 12, 7328 on Dec.16, 2021
(https://doi.org/10.1038/s41467-021-27606-9)
-ProfileProfessor Se-Bum PaikVisual System and Neural Network LaboratoryProgram of Brain and Cognitive EngineeringDepartment of Bio and Brain EngineeringCollege of EngineeringKAIST
2021.12.21 View 11198
Face Detection in Untrained Deep Neural Networks
A KAIST team shows that primitive visual selectivity of faces can arise spontaneously in completely untrained deep neural networks
Researchers have found that higher visual cognitive functions can arise spontaneously in untrained neural networks. A KAIST research team led by Professor Se-Bum Paik from the Department of Bio and Brain Engineering has shown that visual selectivity of facial images can arise even in completely untrained deep neural networks.
This new finding has provided revelatory insights into mechanisms underlying the development of cognitive functions in both biological and artificial neural networks, also making a significant impact on our understanding of the origin of early brain functions before sensory experiences.
The study published in Nature Communications on December 16 demonstrates that neuronal activities selective to facial images are observed in randomly initialized deep neural networks in the complete absence of learning, and that they show the characteristics of those observed in biological brains.
The ability to identify and recognize faces is a crucial function for social behavior, and this ability is thought to originate from neuronal tuning at the single or multi-neuronal level. Neurons that selectively respond to faces are observed in young animals of various species, and this raises intense debate whether face-selective neurons can arise innately in the brain or if they require visual experience.
Using a model neural network that captures properties of the ventral stream of the visual cortex, the research team found that face-selectivity can emerge spontaneously from random feedforward wirings in untrained deep neural networks. The team showed that the character of this innate face-selectivity is comparable to that observed with face-selective neurons in the brain, and that this spontaneous neuronal tuning for faces enables the network to perform face detection tasks.
These results imply a possible scenario in which the random feedforward connections that develop in early, untrained networks may be sufficient for initializing primitive visual cognitive functions.
Professor Paik said, “Our findings suggest that innate cognitive functions can emerge spontaneously from the statistical complexity embedded in the hierarchical feedforward projection circuitry, even in the complete absence of learning”.
He continued, “Our results provide a broad conceptual advance as well as advanced insight into the mechanisms underlying the development of innate functions in both biological and artificial neural networks, which may unravel the mystery of the generation and evolution of intelligence.” This work was supported by the National Research Foundation of Korea (NRF) and by the KAIST singularity research project.
-PublicationSeungdae Baek, Min Song, Jaeson Jang, Gwangsu Kim, and Se-Bum Baik, “Face detection in untrained deep neural network,” Nature Communications 12, 7328 on Dec.16, 2021
(https://doi.org/10.1038/s41467-021-27606-9)
-ProfileProfessor Se-Bum PaikVisual System and Neural Network LaboratoryProgram of Brain and Cognitive EngineeringDepartment of Bio and Brain EngineeringCollege of EngineeringKAIST
2021.12.21 View 11198 -
 Deep Learning Framework to Enable Material Design in Unseen Domain
Researchers propose a deep neural network-based forward design space exploration using active transfer learning and data augmentation
A new study proposed a deep neural network-based forward design approach that enables an efficient search for superior materials far beyond the domain of the initial training set. This approach compensates for the weak predictive power of neural networks on an unseen domain through gradual updates of the neural network with active transfer learning and data augmentation methods.
Professor Seungwha Ryu believes that this study will help address a variety of optimization problems that have an astronomical number of possible design configurations. For the grid composite optimization problem, the proposed framework was able to provide excellent designs close to the global optima, even with the addition of a very small dataset corresponding to less than 0.5% of the initial training data-set size. This study was reported in npj Computational Materials last month.
“We wanted to mitigate the limitation of the neural network, weak predictive power beyond the training set domain for the material or structure design,” said Professor Ryu from the Department of Mechanical Engineering.
Neural network-based generative models have been actively investigated as an inverse design method for finding novel materials in a vast design space. However, the applicability of conventional generative models is limited because they cannot access data outside the range of training sets. Advanced generative models that were devised to overcome this limitation also suffer from weak predictive power for the unseen domain.
Professor Ryu’s team, in collaboration with researchers from Professor Grace Gu’s group at UC Berkeley, devised a design method that simultaneously expands the domain using the strong predictive power of a deep neural network and searches for the optimal design by repetitively performing three key steps.
First, it searches for few candidates with improved properties located close to the training set via genetic algorithms, by mixing superior designs within the training set. Then, it checks to see if the candidates really have improved properties, and expands the training set by duplicating the validated designs via a data augmentation method. Finally, they can expand the reliable prediction domain by updating the neural network with the new superior designs via transfer learning. Because the expansion proceeds along relatively narrow but correct routes toward the optimal design (depicted in the schematic of Fig. 1), the framework enables an efficient search.
As a data-hungry method, a deep neural network model tends to have reliable predictive power only within and near the domain of the training set. When the optimal configuration of materials and structures lies far beyond the initial training set, which frequently is the case, neural network-based design methods suffer from weak predictive power and become inefficient.
Researchers expect that the framework will be applicable for a wide range of optimization problems in other science and engineering disciplines with astronomically large design space, because it provides an efficient way of gradually expanding the reliable prediction domain toward the target design while avoiding the risk of being stuck in local minima. Especially, being a less-data-hungry method, design problems in which data generation is time-consuming and expensive will benefit most from this new framework.
The research team is currently applying the optimization framework for the design task of metamaterial structures, segmented thermoelectric generators, and optimal sensor distributions. “From these sets of on-going studies, we expect to better recognize the pros and cons, and the potential of the suggested algorithm. Ultimately, we want to devise more efficient machine learning-based design approaches,” explained Professor Ryu.This study was funded by the National Research Foundation of Korea and the KAIST Global Singularity Research Project.
-Publication
Yongtae Kim, Youngsoo, Charles Yang, Kundo Park, Grace X. Gu, and Seunghwa Ryu, “Deep learning framework for material design space exploration using active transfer learning and data augmentation,” npj Computational Materials (https://doi.org/10.1038/s41524-021-00609-2)
-Profile
Professor Seunghwa Ryu
Mechanics & Materials Modeling Lab
Department of Mechanical Engineering
KAIST
2021.09.29 View 13289
Deep Learning Framework to Enable Material Design in Unseen Domain
Researchers propose a deep neural network-based forward design space exploration using active transfer learning and data augmentation
A new study proposed a deep neural network-based forward design approach that enables an efficient search for superior materials far beyond the domain of the initial training set. This approach compensates for the weak predictive power of neural networks on an unseen domain through gradual updates of the neural network with active transfer learning and data augmentation methods.
Professor Seungwha Ryu believes that this study will help address a variety of optimization problems that have an astronomical number of possible design configurations. For the grid composite optimization problem, the proposed framework was able to provide excellent designs close to the global optima, even with the addition of a very small dataset corresponding to less than 0.5% of the initial training data-set size. This study was reported in npj Computational Materials last month.
“We wanted to mitigate the limitation of the neural network, weak predictive power beyond the training set domain for the material or structure design,” said Professor Ryu from the Department of Mechanical Engineering.
Neural network-based generative models have been actively investigated as an inverse design method for finding novel materials in a vast design space. However, the applicability of conventional generative models is limited because they cannot access data outside the range of training sets. Advanced generative models that were devised to overcome this limitation also suffer from weak predictive power for the unseen domain.
Professor Ryu’s team, in collaboration with researchers from Professor Grace Gu’s group at UC Berkeley, devised a design method that simultaneously expands the domain using the strong predictive power of a deep neural network and searches for the optimal design by repetitively performing three key steps.
First, it searches for few candidates with improved properties located close to the training set via genetic algorithms, by mixing superior designs within the training set. Then, it checks to see if the candidates really have improved properties, and expands the training set by duplicating the validated designs via a data augmentation method. Finally, they can expand the reliable prediction domain by updating the neural network with the new superior designs via transfer learning. Because the expansion proceeds along relatively narrow but correct routes toward the optimal design (depicted in the schematic of Fig. 1), the framework enables an efficient search.
As a data-hungry method, a deep neural network model tends to have reliable predictive power only within and near the domain of the training set. When the optimal configuration of materials and structures lies far beyond the initial training set, which frequently is the case, neural network-based design methods suffer from weak predictive power and become inefficient.
Researchers expect that the framework will be applicable for a wide range of optimization problems in other science and engineering disciplines with astronomically large design space, because it provides an efficient way of gradually expanding the reliable prediction domain toward the target design while avoiding the risk of being stuck in local minima. Especially, being a less-data-hungry method, design problems in which data generation is time-consuming and expensive will benefit most from this new framework.
The research team is currently applying the optimization framework for the design task of metamaterial structures, segmented thermoelectric generators, and optimal sensor distributions. “From these sets of on-going studies, we expect to better recognize the pros and cons, and the potential of the suggested algorithm. Ultimately, we want to devise more efficient machine learning-based design approaches,” explained Professor Ryu.This study was funded by the National Research Foundation of Korea and the KAIST Global Singularity Research Project.
-Publication
Yongtae Kim, Youngsoo, Charles Yang, Kundo Park, Grace X. Gu, and Seunghwa Ryu, “Deep learning framework for material design space exploration using active transfer learning and data augmentation,” npj Computational Materials (https://doi.org/10.1038/s41524-021-00609-2)
-Profile
Professor Seunghwa Ryu
Mechanics & Materials Modeling Lab
Department of Mechanical Engineering
KAIST
2021.09.29 View 13289 -
 Prof. Sang Wan Lee Selected for 2021 IBM Academic Award
Professor Sang Wan Lee from the Department of Bio and Brain Engineering was selected as the recipient of the 2021 IBM Global University Program Academic Award. The award recognizes individual faculty members whose emerging science and technology contains significant interest for universities and IBM.
Professor Lee, whose research focuses on artificial intelligence and computational neuroscience, won the award for his research proposal titled A Neuroscience-Inspired Approach for Metacognitive Reinforcement Learning. IBM provides a gift of $40,000 to the recipient’s institution in recognition of the selection of the project but not as a contract for services.
Professor Lee’s project aims to exploit the unique characteristics of human reinforcement learning. Specifically, he plans to examines the hypothesis that metacognition, a human’s ability to estimate their uncertainty level, serves to guide sample-efficient and near-optimal exploration, making it possible to achieve an optimal balance between model-based and model-free reinforcement learning.
He was also selected as the winner of the Google Research Award in 2016 and has been working with DeepMind and University College London to conduct basic research on decision-making brain science to establish a theory on frontal lobe meta-enhance learning.
"We plan to conduct joint research for utilizing brain-based artificial intelligence technology and frontal lobe meta-enhanced learning technology modeling in collaboration with an international research team including IBM, DeepMind, MIT, and Oxford,” Professor Lee said.
2021.06.25 View 13683
Prof. Sang Wan Lee Selected for 2021 IBM Academic Award
Professor Sang Wan Lee from the Department of Bio and Brain Engineering was selected as the recipient of the 2021 IBM Global University Program Academic Award. The award recognizes individual faculty members whose emerging science and technology contains significant interest for universities and IBM.
Professor Lee, whose research focuses on artificial intelligence and computational neuroscience, won the award for his research proposal titled A Neuroscience-Inspired Approach for Metacognitive Reinforcement Learning. IBM provides a gift of $40,000 to the recipient’s institution in recognition of the selection of the project but not as a contract for services.
Professor Lee’s project aims to exploit the unique characteristics of human reinforcement learning. Specifically, he plans to examines the hypothesis that metacognition, a human’s ability to estimate their uncertainty level, serves to guide sample-efficient and near-optimal exploration, making it possible to achieve an optimal balance between model-based and model-free reinforcement learning.
He was also selected as the winner of the Google Research Award in 2016 and has been working with DeepMind and University College London to conduct basic research on decision-making brain science to establish a theory on frontal lobe meta-enhance learning.
"We plan to conduct joint research for utilizing brain-based artificial intelligence technology and frontal lobe meta-enhanced learning technology modeling in collaboration with an international research team including IBM, DeepMind, MIT, and Oxford,” Professor Lee said.
2021.06.25 View 13683 -
 Professor Mu-Hyun Baik Honored with the POSCO TJ Park Prize
Professor Mu-Hyun Baik at the Department of Chemistry was honored to be the recipient of the 2021 POSCO TJ Park Prize in Science. The POSCO TJ Park Foundation awards every year the individual or organization which made significant contribution in science, education, community development, philanthropy, and technology.
Professor Baik, a renowned computational chemist in analyzing complicated chemical reactions to understand how molecules behave and how they change. Professor Baik was awarded in recognition of his pioneering research in designing numerous organometallic catalysts with using computational molecular modelling. In 2016, he published in Science on the catalytic borylation of methane that showed how chemical reactions can be carried out using the natural gas methane as a substrate.
In 2020, he reported in Science that electrodes can be used as functional groups with adjustable inductive effects to change the chemical reactivity of molecules that are attached to them, closely mimicking the inductive effect of conventional functional groups. This constitutes a potentially powerful new way of controlling chemical reactions, offering an alternative to preparing derivatives to install electron-withdrawing functional groups.
Joined at KAIST in 2015, Professor Baik also serves as associate director at the Center for Catalytic Hydrocarbon Functionalization at the Institute for Basic Science (IBS) since 2015. Among the many recognitions and awards that he received include the Kavli Fellowship by the Kavli Foundation and the National Academy of Science in the US in 2019 and the 2018 Friedrich Wilhelm Bessel Award by the Alexander von Humboldt Foundation in Germany.
2021.03.11 View 10001
Professor Mu-Hyun Baik Honored with the POSCO TJ Park Prize
Professor Mu-Hyun Baik at the Department of Chemistry was honored to be the recipient of the 2021 POSCO TJ Park Prize in Science. The POSCO TJ Park Foundation awards every year the individual or organization which made significant contribution in science, education, community development, philanthropy, and technology.
Professor Baik, a renowned computational chemist in analyzing complicated chemical reactions to understand how molecules behave and how they change. Professor Baik was awarded in recognition of his pioneering research in designing numerous organometallic catalysts with using computational molecular modelling. In 2016, he published in Science on the catalytic borylation of methane that showed how chemical reactions can be carried out using the natural gas methane as a substrate.
In 2020, he reported in Science that electrodes can be used as functional groups with adjustable inductive effects to change the chemical reactivity of molecules that are attached to them, closely mimicking the inductive effect of conventional functional groups. This constitutes a potentially powerful new way of controlling chemical reactions, offering an alternative to preparing derivatives to install electron-withdrawing functional groups.
Joined at KAIST in 2015, Professor Baik also serves as associate director at the Center for Catalytic Hydrocarbon Functionalization at the Institute for Basic Science (IBS) since 2015. Among the many recognitions and awards that he received include the Kavli Fellowship by the Kavli Foundation and the National Academy of Science in the US in 2019 and the 2018 Friedrich Wilhelm Bessel Award by the Alexander von Humboldt Foundation in Germany.
2021.03.11 View 10001 -
 Sungjoon Park Named Google PhD Fellow
PhD candidate Sungjoon Park from the School of Computing was named a 2019 Google PhD Fellow in the field of natural language processing. The Google PhD fellowship program has recognized and supported outstanding graduate students in computer science and related fields since 2009. Park is one of three Korean students chosen as the recipients of Google Fellowships this year. A total of 54 students across the world in 12 fields were awarded this fellowship.
Park’s research on computational psychotherapy using natural language processing (NLP) powered by machine learning earned him this year’s fellowship. He presented of learning distributed representations in Korean and their interpretations during the 2017 Annual Conference of the Association for Computational Linguistics and the 2018 Conference on Empirical Methods in Natural Language Processing. He also applied machine learning-based natural language processing into computational psychotherapy so that a trained machine learning model could categorize client's verbal responses in a counseling dialogue. This was presented at the Annual Conference of the North American Chapter of the Association for Computational Linguistics.
More recently, he has been developing on neural response generation model and the prediction and extraction of complex emotion in text, and computational psychotherapy applications.
2019.09.17 View 10241
Sungjoon Park Named Google PhD Fellow
PhD candidate Sungjoon Park from the School of Computing was named a 2019 Google PhD Fellow in the field of natural language processing. The Google PhD fellowship program has recognized and supported outstanding graduate students in computer science and related fields since 2009. Park is one of three Korean students chosen as the recipients of Google Fellowships this year. A total of 54 students across the world in 12 fields were awarded this fellowship.
Park’s research on computational psychotherapy using natural language processing (NLP) powered by machine learning earned him this year’s fellowship. He presented of learning distributed representations in Korean and their interpretations during the 2017 Annual Conference of the Association for Computational Linguistics and the 2018 Conference on Empirical Methods in Natural Language Processing. He also applied machine learning-based natural language processing into computational psychotherapy so that a trained machine learning model could categorize client's verbal responses in a counseling dialogue. This was presented at the Annual Conference of the North American Chapter of the Association for Computational Linguistics.
More recently, he has been developing on neural response generation model and the prediction and extraction of complex emotion in text, and computational psychotherapy applications.
2019.09.17 View 10241 -
 Algorithm Identifies Optimal Pairs for Composing Metal-Organic Frameworks
The integration of metal-organic frameworks (MOFs) and other metal nanoparticles has increasingly led to the creation of new multifunctional materials. Many researchers have integrated MOFs with other classes of materials to produce new structures with synergetic properties.
Despite there being over 70,000 collections of synthesized MOFs that can be used as building blocks, the precise nature of the interaction and the bonding at the interface between the two materials still remains unknown. The question is how to sort out the right matching pairs out of 70,000 MOFs.
An algorithmic study published in Nature Communications by a KAIST research team presents a clue for finding the perfect pairs. The team, led by Professor Ji-Han Kim from the Department of Chemical and Biomolecular Engineering, developed a joint computational and experimental approach to rationally design MOF@MOFs, a composite of MOFs where an MOF is grown on a different MOF.
Professor Kim’s team, in collaboration with UNIST, noted that the metal node of one MOF can coordinately bond with the linker of a different MOF and the precisely matched interface configurations at atomic and molecular levels can enhance the likelihood of synthesizing MOF@MOFs.
They screened thousands of MOFs and identified optimal MOF pairs that can seamlessly connect to one another by taking advantage of the fact that the metal node of one MOF can form coordination bonds with the linkers of the second MOF. Six pairs predicted from the computational algorithm successfully grew into single crystals.
This computational workflow can readily extend into other classes of materials and can lead to the rapid exploration of the composite MOFs arena for accelerated materials development. Even more, the workflow can enhance the likelihood of synthesizing MOF@MOFs in the form of large single crystals, and thereby demonstrated the utility of rationally designing the MOF@MOFs.
This study is the first algorithm for predicting the synthesis of composite MOFs, to the best of their knowledge. Professor Kim said, “The number of predicted pairs can increase even more with the more general 2D lattice matching, and it is worth investigating in the future.”
This study was supported by Samsung Research Funding & Incubation Center of Samsung Electronics.
(Figure: An example of a rationally synthesized MOF@MOFs (cubic HKUST-1@MOF-5 ))
2019.08.30 View 17861
Algorithm Identifies Optimal Pairs for Composing Metal-Organic Frameworks
The integration of metal-organic frameworks (MOFs) and other metal nanoparticles has increasingly led to the creation of new multifunctional materials. Many researchers have integrated MOFs with other classes of materials to produce new structures with synergetic properties.
Despite there being over 70,000 collections of synthesized MOFs that can be used as building blocks, the precise nature of the interaction and the bonding at the interface between the two materials still remains unknown. The question is how to sort out the right matching pairs out of 70,000 MOFs.
An algorithmic study published in Nature Communications by a KAIST research team presents a clue for finding the perfect pairs. The team, led by Professor Ji-Han Kim from the Department of Chemical and Biomolecular Engineering, developed a joint computational and experimental approach to rationally design MOF@MOFs, a composite of MOFs where an MOF is grown on a different MOF.
Professor Kim’s team, in collaboration with UNIST, noted that the metal node of one MOF can coordinately bond with the linker of a different MOF and the precisely matched interface configurations at atomic and molecular levels can enhance the likelihood of synthesizing MOF@MOFs.
They screened thousands of MOFs and identified optimal MOF pairs that can seamlessly connect to one another by taking advantage of the fact that the metal node of one MOF can form coordination bonds with the linkers of the second MOF. Six pairs predicted from the computational algorithm successfully grew into single crystals.
This computational workflow can readily extend into other classes of materials and can lead to the rapid exploration of the composite MOFs arena for accelerated materials development. Even more, the workflow can enhance the likelihood of synthesizing MOF@MOFs in the form of large single crystals, and thereby demonstrated the utility of rationally designing the MOF@MOFs.
This study is the first algorithm for predicting the synthesis of composite MOFs, to the best of their knowledge. Professor Kim said, “The number of predicted pairs can increase even more with the more general 2D lattice matching, and it is worth investigating in the future.”
This study was supported by Samsung Research Funding & Incubation Center of Samsung Electronics.
(Figure: An example of a rationally synthesized MOF@MOFs (cubic HKUST-1@MOF-5 ))
2019.08.30 View 17861 -
 Professor Ji-Hyun Lee Awarded the Sasada Prize
Professor Ji-Hyun Lee from the Graduate School of Culture Technology was awarded the Sasada Prize during the 24th annual Conference of Computer-Aided Architectural Design Research in Asia (CAADRIA) held in Wellington, New Zealand on April 15.
The Sasada Award honors the late Professor Tsuyoshi Sasada (1941-2005), the former Professor of Osaka University and co-founder and fellow of CAADRIA. It is given to an individual who has contributed to the next generation of researchers and academics, to the wider profession and practice in computer-aided design and research, and has earned recognition in the academic community.
Professor Lee was recognized for her development of CAAD (Computer-Aided Architectural Design) through her research work on the land price precision system using case-based reasoning. Her research team proposed a model for estimating the average apartment price in an administrative district after collecting 40 variables from the six major Korean cities, excluding Seoul and Ulsan. Their follow-up studies showed the possibility of replacing existing experts’ predictions.
Professor Lee has been steadily researching for 20 years on case-based reasoning (CBR), a field of artificial intelligence, and has published more than 40 papers in the field of CBR. Meanwhile, the CAAD Future 2019 event will be held at KAIST in June.
2019.04.23 View 8054
Professor Ji-Hyun Lee Awarded the Sasada Prize
Professor Ji-Hyun Lee from the Graduate School of Culture Technology was awarded the Sasada Prize during the 24th annual Conference of Computer-Aided Architectural Design Research in Asia (CAADRIA) held in Wellington, New Zealand on April 15.
The Sasada Award honors the late Professor Tsuyoshi Sasada (1941-2005), the former Professor of Osaka University and co-founder and fellow of CAADRIA. It is given to an individual who has contributed to the next generation of researchers and academics, to the wider profession and practice in computer-aided design and research, and has earned recognition in the academic community.
Professor Lee was recognized for her development of CAAD (Computer-Aided Architectural Design) through her research work on the land price precision system using case-based reasoning. Her research team proposed a model for estimating the average apartment price in an administrative district after collecting 40 variables from the six major Korean cities, excluding Seoul and Ulsan. Their follow-up studies showed the possibility of replacing existing experts’ predictions.
Professor Lee has been steadily researching for 20 years on case-based reasoning (CBR), a field of artificial intelligence, and has published more than 40 papers in the field of CBR. Meanwhile, the CAAD Future 2019 event will be held at KAIST in June.
2019.04.23 View 8054