Biomolecular+Engineering+Lab
-
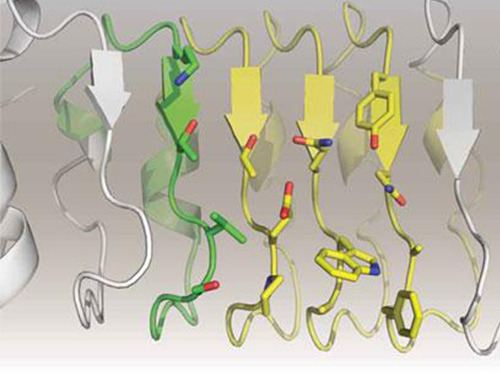 Artificial Antibody-based Therapeutic Candidate for Lung Cancer Developed
Professor Hak-Sung Kim of Biological Sciences at KAIST publishes a cover article on artificial antibody in "Molecular Therapy".
Repebody-based lung cancer therapeutic drug candidate developed Repebody-based protein demonstrates the possibility of the development of a new drug
KAIST Biological Sciences Department’s Professor Hak-Sung Kim, in collaboration with Professor Eun-Kyung Cho from the College of Medicine at Chungnam National University, has successfully developed an artificial antibody-based, or repebody, cancer therapeutic candidate. These research results were published as a cover paper of the July edition of Molecular Therapy.
The repebody developed by Professor Kim and his team strongly binds to interleukin-6, a cancer-causing factor. It has also been confirmed that the repebody can significantly inhibit the proliferation of cancer cells in non-small-cell lung cancer animal model.
Numerous multinational pharmaceutical and biotechnology companies have invested astronomical amounts of money in research for the development of protein therapeutics with low side effects and high efficacy. More than 20 kinds of such therapeutics are currently under clinical trials, and over 100 drugs are under clinical demonstration. Among these, the majority is antibody-based therapeutics, and most of the investments are heavily concentrated in this field. However, antibody production cost is very high because it has large molecular weights and complex structural properties, and this makes it difficult to engineer. Consequently, the development costs a great deal of time and money.
In order to overcome the existing limitations of antibody-based therapeutics, Professor Kim and his team have developed a new artificial antibody, or repebody, which was published in Proceedings of the National Academy of Sciences (PNAS) in 2012. Based on this research, they have succeeded in developing a therapeutic candidate for treating non-small-cell lung cancer with a specifically strong cohesion to the cancer-causing factor, interleukin-6.
Interleukin-6 is a crucial substance within the body that is involved in immune and inflammatory-related signals. When abnormally expressed, it activates various carcinogenic pathways and promotes tumor growth and metastasis. Because of its importance, multinational pharmaceutical companies are heavily investing in developing therapeutics that can inhibit the signaling of interleukin-6.
In this study, Professor Kim and his team observed that a repebody consists of repeated modules, and they conceived a module-based affinity amplification technology that can effectively increase the binding affinity with the disease target. The developed therapeutic candidate has been confirmed in cell and animal experiments to show low immunogenicity, as well as to strongly inhibit the proliferation of non-small-cell lung cancer.
Furthermore, by investigating the complex structure of the repebody with interleukin-6, Professor Kim has identified its mechanism, which demonstrated the potential for therapeutic development. The researchers are currently carrying out pre-clinical trials for acquiring permission to perform clinical trials on animals with non-small-cell lung cancer. The repebody can be developed into a new protein drug after demonstrating its safety and efficacy.
Professor Hak-Sung Kim and his team have confirmed that the repebody can be utilized as a new protein drug, and this will be a significant contribution to Korea’s protein drugs and biotechnology industry development.
The research was supported by the Future Pioneer Industry project and sponsored by the Ministry of Science, ICT and Future Planning.
Figure 1. Professor Kim’s article published as the cover article of July edition of Molecular Therapy
Figure 2. Clinical proof of the repebody’s inhibition of cancer growth using animal models
2014.07.14 View 15103
Artificial Antibody-based Therapeutic Candidate for Lung Cancer Developed
Professor Hak-Sung Kim of Biological Sciences at KAIST publishes a cover article on artificial antibody in "Molecular Therapy".
Repebody-based lung cancer therapeutic drug candidate developed Repebody-based protein demonstrates the possibility of the development of a new drug
KAIST Biological Sciences Department’s Professor Hak-Sung Kim, in collaboration with Professor Eun-Kyung Cho from the College of Medicine at Chungnam National University, has successfully developed an artificial antibody-based, or repebody, cancer therapeutic candidate. These research results were published as a cover paper of the July edition of Molecular Therapy.
The repebody developed by Professor Kim and his team strongly binds to interleukin-6, a cancer-causing factor. It has also been confirmed that the repebody can significantly inhibit the proliferation of cancer cells in non-small-cell lung cancer animal model.
Numerous multinational pharmaceutical and biotechnology companies have invested astronomical amounts of money in research for the development of protein therapeutics with low side effects and high efficacy. More than 20 kinds of such therapeutics are currently under clinical trials, and over 100 drugs are under clinical demonstration. Among these, the majority is antibody-based therapeutics, and most of the investments are heavily concentrated in this field. However, antibody production cost is very high because it has large molecular weights and complex structural properties, and this makes it difficult to engineer. Consequently, the development costs a great deal of time and money.
In order to overcome the existing limitations of antibody-based therapeutics, Professor Kim and his team have developed a new artificial antibody, or repebody, which was published in Proceedings of the National Academy of Sciences (PNAS) in 2012. Based on this research, they have succeeded in developing a therapeutic candidate for treating non-small-cell lung cancer with a specifically strong cohesion to the cancer-causing factor, interleukin-6.
Interleukin-6 is a crucial substance within the body that is involved in immune and inflammatory-related signals. When abnormally expressed, it activates various carcinogenic pathways and promotes tumor growth and metastasis. Because of its importance, multinational pharmaceutical companies are heavily investing in developing therapeutics that can inhibit the signaling of interleukin-6.
In this study, Professor Kim and his team observed that a repebody consists of repeated modules, and they conceived a module-based affinity amplification technology that can effectively increase the binding affinity with the disease target. The developed therapeutic candidate has been confirmed in cell and animal experiments to show low immunogenicity, as well as to strongly inhibit the proliferation of non-small-cell lung cancer.
Furthermore, by investigating the complex structure of the repebody with interleukin-6, Professor Kim has identified its mechanism, which demonstrated the potential for therapeutic development. The researchers are currently carrying out pre-clinical trials for acquiring permission to perform clinical trials on animals with non-small-cell lung cancer. The repebody can be developed into a new protein drug after demonstrating its safety and efficacy.
Professor Hak-Sung Kim and his team have confirmed that the repebody can be utilized as a new protein drug, and this will be a significant contribution to Korea’s protein drugs and biotechnology industry development.
The research was supported by the Future Pioneer Industry project and sponsored by the Ministry of Science, ICT and Future Planning.
Figure 1. Professor Kim’s article published as the cover article of July edition of Molecular Therapy
Figure 2. Clinical proof of the repebody’s inhibition of cancer growth using animal models
2014.07.14 View 15103 -
 Binding Regulatory Mechanism of Protein Biomolecules Revealed
Professor Hak-Sung Kim
A research team led by Professor Hak-Sung Kim of Biological Sciences, KAIST, and Dr. Mun-Hyeong Seo, KAIST, has revealed a regulatory mechanism that controls the binding affinity of protein’s biomolecules, which is crucial for the protein to recognize molecules and carry out functions within the body.
The research results were published in the April 24th online edition of Nature Communications.
The protein, represented by enzyme, antibody, or hormones, specifically recognizes a variety of biomolecules in all organisms and implements signaling or immune response to precisely adjust and maintain important biological processes. The protein binding affinity of biomolecules plays a crucial role in determining the duration of the bond between two molecules, and hence to determine and control the in-vivo function of proteins.
The researchers have noted that, during the process of proteins’ recognizing biomolecules, the protein binding affinity of biomolecules is closely linked not only to the size of non-covalent interaction between two molecules, but also to the unique kinetic properties of proteins.
To identify the basic mechanism that determines the protein binding affinity of biomolecules, Professor Kim and his research team have made mutation in the allosteric site of protein to create a variety of mutant proteins with the same chemical binding surface, but with the binding affinity vastly differing from 10 to 100 times. The allosteric site of the protein refers to a region which does not directly bind with biomolecules, but crucially influences the biomolecule recognition site.
Using real-time analysis at the single-molecule level of unique kinetic properties of the produced mutant proteins, the researchers were able to identify that the protein binding affinity of biomolecules is directly associated with the protein’s specific kinetic characteristics, its structure opening rate.
Also, by proving that unique characteristics of the protein can be changed at the allosteric site, instead of protein’s direct binding site with biomolecules, the researchers have demonstrated a new methodology of regulating the in-vivo function of proteins.
The researchers expect that these results will contribute greatly to a deeper understanding of protein’s nature that governs various life phenomena and help evaluate the proof of interpreting protein binding affinity of biomolecules from the perspective of protein kinetics.
Professor Kim said, “Until now, the protein binding affinity of biomolecules was determined by a direct interaction between two molecules. Our research has identified an important fact that the structure opening rate of proteins also plays a crucial role in determining their binding affinity.”
[Picture]
A correlation graph of opening rate (kopening) and binding affinity (kd) between protein’s stable, open state and its unstable, partially closed state.
2014.05.02 View 11457
Binding Regulatory Mechanism of Protein Biomolecules Revealed
Professor Hak-Sung Kim
A research team led by Professor Hak-Sung Kim of Biological Sciences, KAIST, and Dr. Mun-Hyeong Seo, KAIST, has revealed a regulatory mechanism that controls the binding affinity of protein’s biomolecules, which is crucial for the protein to recognize molecules and carry out functions within the body.
The research results were published in the April 24th online edition of Nature Communications.
The protein, represented by enzyme, antibody, or hormones, specifically recognizes a variety of biomolecules in all organisms and implements signaling or immune response to precisely adjust and maintain important biological processes. The protein binding affinity of biomolecules plays a crucial role in determining the duration of the bond between two molecules, and hence to determine and control the in-vivo function of proteins.
The researchers have noted that, during the process of proteins’ recognizing biomolecules, the protein binding affinity of biomolecules is closely linked not only to the size of non-covalent interaction between two molecules, but also to the unique kinetic properties of proteins.
To identify the basic mechanism that determines the protein binding affinity of biomolecules, Professor Kim and his research team have made mutation in the allosteric site of protein to create a variety of mutant proteins with the same chemical binding surface, but with the binding affinity vastly differing from 10 to 100 times. The allosteric site of the protein refers to a region which does not directly bind with biomolecules, but crucially influences the biomolecule recognition site.
Using real-time analysis at the single-molecule level of unique kinetic properties of the produced mutant proteins, the researchers were able to identify that the protein binding affinity of biomolecules is directly associated with the protein’s specific kinetic characteristics, its structure opening rate.
Also, by proving that unique characteristics of the protein can be changed at the allosteric site, instead of protein’s direct binding site with biomolecules, the researchers have demonstrated a new methodology of regulating the in-vivo function of proteins.
The researchers expect that these results will contribute greatly to a deeper understanding of protein’s nature that governs various life phenomena and help evaluate the proof of interpreting protein binding affinity of biomolecules from the perspective of protein kinetics.
Professor Kim said, “Until now, the protein binding affinity of biomolecules was determined by a direct interaction between two molecules. Our research has identified an important fact that the structure opening rate of proteins also plays a crucial role in determining their binding affinity.”
[Picture]
A correlation graph of opening rate (kopening) and binding affinity (kd) between protein’s stable, open state and its unstable, partially closed state.
2014.05.02 View 11457 -
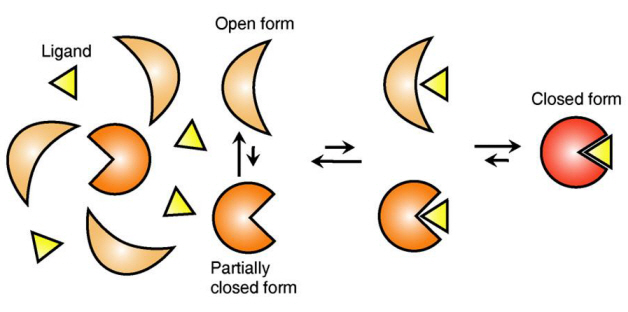 Ligand Recognition Mechanism of Protein Identified
Professor Hak-Sung Kim
-“Solved the 50 year old mystery of how protein recognises and binds to ligands”
- Exciting potential for understanding life phenomena and the further development of highly effective therapeutic agent development
KAIST’s Biological Science Department’s Professor Hak-Sung Kim, working in collaboration with Professor Sung-Chul Hong of Department of Physics, Seoul National University, has identified the mechanism of how the protein recognizes and binds to ligands within the human body.
The research findings were published in the online edition of Nature Chemical Biology (March 18), which is the most prestigious journal in the field of life science.
Since the research identified the mechanism, of which protein recognises and binds to ligands, it will take an essential role in understanding complex life phenomenon by understanding regulatory function of protein.
Also, ligand recognition of proteins is closely related to the cause of various diseases. Therefore the research team hopes to contribute to the development of highly effective treatments.
Ligands, well-known examples include nucleic acid and proteins, form the structure of an organism or are essential constituents with special functions such as information signalling.
In particular, the most important role of protein is recognising and binding to a particular ligand and hence regulating and maintaining life phenomena. The abnormal occurrence of an error in recognition of ligands may lead to various diseases.
The research team focused on the repetition of change in protein structure from the most stable “open form” to a relatively unstable “partially closed form”.
Professor Kim’s team analysed the change in protein structure when binding to a ligand on a molecular level in real time to explain the ligand recognition mechanism.
The research findings showed that ligands prefer the most stable protein structure. The team was the first in the world to identify that ligands alter protein structure to the most stable, the lowest energy level, when it binds to the protein.
In addition, the team found that ligands bind to unstable partially-closed forms to change protein structure.
The existing models to explain ligand recognition mechanism of protein are “Induced Custom Model”, which involves change in protein structure in binding to ligands, and the “Structure Selection Model”, which argues that ligands select and recognise only the best protein structure out of many. The academic world considers that the team’s research findings have perfectly proved the models through experiments for the first time in the world.
Professor Kim explained, “In the presence of ligands, there exists a phenomenon where the speed of altering protein structure is changed. This phenomenon is analysed on a molecular level to prove ligand recognition mechanism of protein for the first time”. He also said, “The 50-year old mystery, that existed only as a hypothesis on biology textbooks and was thought never to be solved, has been confirmed through experiments for the first time.”
Figure 1: Proteins, with open and partially open form, recognising and binding to ligands.
Figure 2: Ligands temporarily bind to a stable protein structure, open form, which changes into the most stable structure, closed form. In addition, binding to partially closed form also changes protein structure to closed form.
2013.04.01 View 14372
Ligand Recognition Mechanism of Protein Identified
Professor Hak-Sung Kim
-“Solved the 50 year old mystery of how protein recognises and binds to ligands”
- Exciting potential for understanding life phenomena and the further development of highly effective therapeutic agent development
KAIST’s Biological Science Department’s Professor Hak-Sung Kim, working in collaboration with Professor Sung-Chul Hong of Department of Physics, Seoul National University, has identified the mechanism of how the protein recognizes and binds to ligands within the human body.
The research findings were published in the online edition of Nature Chemical Biology (March 18), which is the most prestigious journal in the field of life science.
Since the research identified the mechanism, of which protein recognises and binds to ligands, it will take an essential role in understanding complex life phenomenon by understanding regulatory function of protein.
Also, ligand recognition of proteins is closely related to the cause of various diseases. Therefore the research team hopes to contribute to the development of highly effective treatments.
Ligands, well-known examples include nucleic acid and proteins, form the structure of an organism or are essential constituents with special functions such as information signalling.
In particular, the most important role of protein is recognising and binding to a particular ligand and hence regulating and maintaining life phenomena. The abnormal occurrence of an error in recognition of ligands may lead to various diseases.
The research team focused on the repetition of change in protein structure from the most stable “open form” to a relatively unstable “partially closed form”.
Professor Kim’s team analysed the change in protein structure when binding to a ligand on a molecular level in real time to explain the ligand recognition mechanism.
The research findings showed that ligands prefer the most stable protein structure. The team was the first in the world to identify that ligands alter protein structure to the most stable, the lowest energy level, when it binds to the protein.
In addition, the team found that ligands bind to unstable partially-closed forms to change protein structure.
The existing models to explain ligand recognition mechanism of protein are “Induced Custom Model”, which involves change in protein structure in binding to ligands, and the “Structure Selection Model”, which argues that ligands select and recognise only the best protein structure out of many. The academic world considers that the team’s research findings have perfectly proved the models through experiments for the first time in the world.
Professor Kim explained, “In the presence of ligands, there exists a phenomenon where the speed of altering protein structure is changed. This phenomenon is analysed on a molecular level to prove ligand recognition mechanism of protein for the first time”. He also said, “The 50-year old mystery, that existed only as a hypothesis on biology textbooks and was thought never to be solved, has been confirmed through experiments for the first time.”
Figure 1: Proteins, with open and partially open form, recognising and binding to ligands.
Figure 2: Ligands temporarily bind to a stable protein structure, open form, which changes into the most stable structure, closed form. In addition, binding to partially closed form also changes protein structure to closed form.
2013.04.01 View 14372 -
 Creation of Synthetic Antibodies: Professor Hak Seong Kim
Synthetics antibodies which can replace antibodies from humans used as ingredients of medicines have been developed. It can increase the costs to 1/100 of the current costs and is much easier to develop. It is expected that the development period will be shortened from 10 years to 5.
Prof. Hak Seong Kim from the Biology department of KAIST conducted a joint research with Prof. Dong Seob Kim to reconstruct proteins and has succeeded.
The synthetic antibody displays much strength in terms of its productivity, structural formation, and bonding capability, and is thus regarded as an ideal protein. It can replace the antigens that are currently in use. It is expected that Korea will therefore be able to lead the world market for protein medicines which is a 192trillion won industry.
The original antibody has been used for not only treating diseases, but also for various other applications in the fields of medical sciences and biology. However, it is produced through a very complex process involving the incubation of animal cells, and is therefore very expensive. Also, most antibodies are already patented by more developed countries, so a high royalty fee must be paid.
Because of this, many countries including Korea has been concentrating on developing biosimilars copying the antibody medicines for which the patents have already expired. This causes Korea to be behind in the development of antibody protein pharmaceuticals.
Prof. Kim’s research team has focused on the face that the protein existing in some eels are not antibodies but functions as one, and has been successful in developing a synthetic antibody.
The synthetic antibody can be mass produced from the colon bacillus, which allows it to be produced at 1/100 the original cost. It is in a module structure which allows the structuring of the antibody into the desired structure, enabling it to be developed into a protein-based medicine within 5 years.
Together with this, the coherence with the important antigens can be easily controlled, thus allowing for highly effective treatments, less side-effects, high security regarding heat and pH, and the immunogen levels being negligeable. This suggests a very high rate of the antibody being converted into a protein based medication.
The synthetic antibody technology has been tested as a sample for the cure for lung diseases and rheumatism and has been proven to be appropriate. Animal testing will be conducted soon.
Prof Kim said “The original antibodies had a small area allowing the bonding with antibodies, creating barriers for raising bonding strength and structuring. The newly created antibody carries only the strengths and will become a new protein based medicine purely created by Korean technology to replace the antibodies currently used in medications.”
Furthermore, he added that, “The synthesized antibody structuring and designing technology will be widely used in the areas of detecting, diagnosing, and analyzing diseases.”
At the same time, this research result has been published in the Feb 10th issue of the PNAS, and has been supported by the future promising pioneer business program held by the Ministry of Education and Technology.
2012.04.04 View 13272
Creation of Synthetic Antibodies: Professor Hak Seong Kim
Synthetics antibodies which can replace antibodies from humans used as ingredients of medicines have been developed. It can increase the costs to 1/100 of the current costs and is much easier to develop. It is expected that the development period will be shortened from 10 years to 5.
Prof. Hak Seong Kim from the Biology department of KAIST conducted a joint research with Prof. Dong Seob Kim to reconstruct proteins and has succeeded.
The synthetic antibody displays much strength in terms of its productivity, structural formation, and bonding capability, and is thus regarded as an ideal protein. It can replace the antigens that are currently in use. It is expected that Korea will therefore be able to lead the world market for protein medicines which is a 192trillion won industry.
The original antibody has been used for not only treating diseases, but also for various other applications in the fields of medical sciences and biology. However, it is produced through a very complex process involving the incubation of animal cells, and is therefore very expensive. Also, most antibodies are already patented by more developed countries, so a high royalty fee must be paid.
Because of this, many countries including Korea has been concentrating on developing biosimilars copying the antibody medicines for which the patents have already expired. This causes Korea to be behind in the development of antibody protein pharmaceuticals.
Prof. Kim’s research team has focused on the face that the protein existing in some eels are not antibodies but functions as one, and has been successful in developing a synthetic antibody.
The synthetic antibody can be mass produced from the colon bacillus, which allows it to be produced at 1/100 the original cost. It is in a module structure which allows the structuring of the antibody into the desired structure, enabling it to be developed into a protein-based medicine within 5 years.
Together with this, the coherence with the important antigens can be easily controlled, thus allowing for highly effective treatments, less side-effects, high security regarding heat and pH, and the immunogen levels being negligeable. This suggests a very high rate of the antibody being converted into a protein based medication.
The synthetic antibody technology has been tested as a sample for the cure for lung diseases and rheumatism and has been proven to be appropriate. Animal testing will be conducted soon.
Prof Kim said “The original antibodies had a small area allowing the bonding with antibodies, creating barriers for raising bonding strength and structuring. The newly created antibody carries only the strengths and will become a new protein based medicine purely created by Korean technology to replace the antibodies currently used in medications.”
Furthermore, he added that, “The synthesized antibody structuring and designing technology will be widely used in the areas of detecting, diagnosing, and analyzing diseases.”
At the same time, this research result has been published in the Feb 10th issue of the PNAS, and has been supported by the future promising pioneer business program held by the Ministry of Education and Technology.
2012.04.04 View 13272