Proceedings+of+the+National+Academy+of+Science
-
 KAIST Researchers Suggest an Extraordinary Alternative to Petroleum-based PET - Bacteria!
< (From left) Dr. Cindy Pricilia, Ph.D. Candidate Cheon Woo Moon, Distinguished Professor Sang Yup Lee >
Currently, the world is suffering from environmental problems caused by plastic waste. The KAIST research team has succeeded in producing a microbial-based plastic that is biodegradable and can replace existing PET bottles, making it a hot topic.
Our university announced on the 7th of November that the research team of Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering has succeeded in developing a microbial strain that efficiently produces pseudoaromatic polyester monomer to replace polyethylene terephthalate (PET) using systems metabolic engineering.
Pseudoaromatic dicarboxylic acids have better physical properties and higher biodegradability than aromatic polyester (PET) when synthesized as polymers, and are attracting attention as an eco-friendly monomer* that can be synthesized into polymers. The production of pseudoaromatic dicarboxylic acids through chemical methods has the problems of low yield and selectivity, complex reaction conditions, and the generation of hazardous waste.
*Monomer: A material for making polymers, which is used to synthesize polymers by polymerizing monomers together
< Figure. Overview of pseudoaromatic dicarboxylic acid production using metabolically engineered C. glutamicum. >
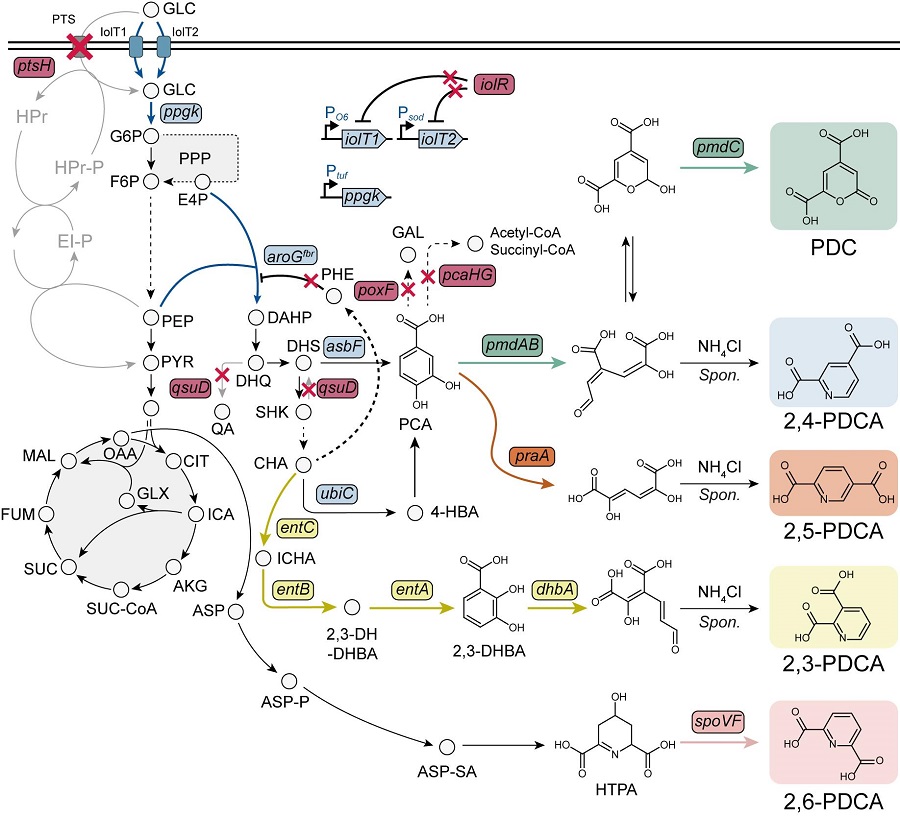
To solve this problem, Professor Sang Yup Lee's research team used metabolic engineering to develop a microbial strain that efficiently produces five types of pseudoaromatic dicarboxylic acids, including 2-pyrone-4,6-dicarboxylic acid and four types of pyridine dicarboxylic acids (2,3-, 2,4-, 2,5-, 2,6-pyridine dicarboxylic acids), in Corynebacterium, a bacterium mainly used for amino acid production.
The research team used metabolic engineering techniques to build a platform microbial strain that enhances the metabolic flow of protocatechuic acid, which is used as a precursor for several pseudoaromatic dicarboxylic acids, and prevents the loss of precursors.
Based on this, the genetic manipulation target was discovered through transcriptome analysis, producing 76.17 g/L of 2-pyrone-4,6-dicarboxylic acid, and by newly discovering and constructing three types of pyridine dicarboxylic acid production metabolic pathways, successfully producing 2.79 g/L of 2,3-pyridine dicarboxylic acid, 0.49 g/L of 2,4-pyridine dicarboxylic acid, and 1.42 g/L of 2,5-pyridine dicarboxylic acid.
In addition, the research team confirmed the production of 15.01 g/L through the construction and reinforcement of the 2,6-pyridine dicarboxylic acid biosynthesis pathway, successfully producing a total of five similar aromatic dicarboxylic acids with high efficiency.
In conclusion, the team succeeded in producing 2,4-, 2,5-, and 2,6-pyridine dicarboxylic acids at the world's highest concentration. In particular, 2,4-, 2,5-pyridine dicarboxylic acid achieved production on the scale of g/L, which was previously produced in extremely small amounts (mg/L).
Based on this study, it is expected that it will be applied to various polyester production industrial processes, and it is also expected that it will be actively utilized in research on the production of similar aromatic polyesters.
Professor Sang Yup Lee, the corresponding author, said, “The significance lies in the fact that we have developed an eco-friendly technology that efficiently produces similar aromatic polyester monomers based on microorganisms,” and “This study will help the microorganism-based bio-monomer industry replace the petrochemical-based chemical industry in the future.”
The results of this study were published in the international academic journal, the Proceedings of the National Academy of Sciences of United States of America (PNAS) on October 30th.
※ Paper title: Metabolic engineering of Corynebacterium glutamicum for the production of pyrone and pyridine dicarboxylic acids
※ Author information: Jae Sung Cho (co-first author), Zi Wei Luo (co-first author), Cheon Woo Moon (co-first author), Cindy Prabowo (co-author), Sang Yup Lee (corresponding author) - a total of 5 people
This study was conducted with the support of the Development of Next-generation Biorefinery Platform Technologies for Leading Bio-based Chemicals Industry Project and the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project (Project leader: Professor Sang Yup Lee) from the National Research Foundation supported by the Ministry of Science and Technology and ICT of Korea.
2024.11.08 View 7760
KAIST Researchers Suggest an Extraordinary Alternative to Petroleum-based PET - Bacteria!
< (From left) Dr. Cindy Pricilia, Ph.D. Candidate Cheon Woo Moon, Distinguished Professor Sang Yup Lee >
Currently, the world is suffering from environmental problems caused by plastic waste. The KAIST research team has succeeded in producing a microbial-based plastic that is biodegradable and can replace existing PET bottles, making it a hot topic.
Our university announced on the 7th of November that the research team of Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering has succeeded in developing a microbial strain that efficiently produces pseudoaromatic polyester monomer to replace polyethylene terephthalate (PET) using systems metabolic engineering.
Pseudoaromatic dicarboxylic acids have better physical properties and higher biodegradability than aromatic polyester (PET) when synthesized as polymers, and are attracting attention as an eco-friendly monomer* that can be synthesized into polymers. The production of pseudoaromatic dicarboxylic acids through chemical methods has the problems of low yield and selectivity, complex reaction conditions, and the generation of hazardous waste.
*Monomer: A material for making polymers, which is used to synthesize polymers by polymerizing monomers together
< Figure. Overview of pseudoaromatic dicarboxylic acid production using metabolically engineered C. glutamicum. >
To solve this problem, Professor Sang Yup Lee's research team used metabolic engineering to develop a microbial strain that efficiently produces five types of pseudoaromatic dicarboxylic acids, including 2-pyrone-4,6-dicarboxylic acid and four types of pyridine dicarboxylic acids (2,3-, 2,4-, 2,5-, 2,6-pyridine dicarboxylic acids), in Corynebacterium, a bacterium mainly used for amino acid production.
The research team used metabolic engineering techniques to build a platform microbial strain that enhances the metabolic flow of protocatechuic acid, which is used as a precursor for several pseudoaromatic dicarboxylic acids, and prevents the loss of precursors.
Based on this, the genetic manipulation target was discovered through transcriptome analysis, producing 76.17 g/L of 2-pyrone-4,6-dicarboxylic acid, and by newly discovering and constructing three types of pyridine dicarboxylic acid production metabolic pathways, successfully producing 2.79 g/L of 2,3-pyridine dicarboxylic acid, 0.49 g/L of 2,4-pyridine dicarboxylic acid, and 1.42 g/L of 2,5-pyridine dicarboxylic acid.
In addition, the research team confirmed the production of 15.01 g/L through the construction and reinforcement of the 2,6-pyridine dicarboxylic acid biosynthesis pathway, successfully producing a total of five similar aromatic dicarboxylic acids with high efficiency.
In conclusion, the team succeeded in producing 2,4-, 2,5-, and 2,6-pyridine dicarboxylic acids at the world's highest concentration. In particular, 2,4-, 2,5-pyridine dicarboxylic acid achieved production on the scale of g/L, which was previously produced in extremely small amounts (mg/L).
Based on this study, it is expected that it will be applied to various polyester production industrial processes, and it is also expected that it will be actively utilized in research on the production of similar aromatic polyesters.
Professor Sang Yup Lee, the corresponding author, said, “The significance lies in the fact that we have developed an eco-friendly technology that efficiently produces similar aromatic polyester monomers based on microorganisms,” and “This study will help the microorganism-based bio-monomer industry replace the petrochemical-based chemical industry in the future.”
The results of this study were published in the international academic journal, the Proceedings of the National Academy of Sciences of United States of America (PNAS) on October 30th.
※ Paper title: Metabolic engineering of Corynebacterium glutamicum for the production of pyrone and pyridine dicarboxylic acids
※ Author information: Jae Sung Cho (co-first author), Zi Wei Luo (co-first author), Cheon Woo Moon (co-first author), Cindy Prabowo (co-author), Sang Yup Lee (corresponding author) - a total of 5 people
This study was conducted with the support of the Development of Next-generation Biorefinery Platform Technologies for Leading Bio-based Chemicals Industry Project and the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project (Project leader: Professor Sang Yup Lee) from the National Research Foundation supported by the Ministry of Science and Technology and ICT of Korea.
2024.11.08 View 7760 -
 Mystery Solved with Math: Cytoplasmic Traffic Jam Disrupts Sleep-Wake Cycles
KAIST mathematicians and their collaborators at Florida State University have identified the principle of how aging and diseases like dementia and obesity cause sleep disorders. A combination of mathematical modelling and experiments demonstrated that the cytoplasmic congestion caused by aging, dementia, and/or obesity disrupts the circadian rhythms in the human body and leads to irregular sleep-wake cycles. This finding suggests new treatment strategies for addressing unstable sleep-wake cycles.
Human bodies adjust sleep schedules in accordance with the ‘circadian rhythms’, which are regulated by our time keeping system, the ‘circadian clock’. This clock tells our body when to rest by generating the 24-hour rhythms of a protein called PERIOD (PER) (See Figure 1).
The amount of the PER protein increases for half of the day and then decreases for the remaining half. The principle is that the PER protein accumulating in the cytoplasm for several hours enters the cell nucleus all at once, hindering the transcription of PER genes and thereby reducing the amount of PER.
However, it has remained a mystery how thousands of PER molecules can simultaneously enter into the nucleus in a complex cell environment where a variety of materials co-exist and can interfere with the motion of PER. This would be like finding a way for thousands of employees from all over New York City to enter an office building at the same time every day.
A group of researchers led by Professor Jae Kyoung Kim from the KAIST Department of Mathematical Sciences solved the mystery by developing a spatiotemporal and probabilistic model that describes the motion of PER molecules in a cell environment.
This study was conducted in collaboration with Professor Choogon Lee’s group from Florida State University, where the experiments were carried out, and the results were published in the Proceedings of the National Academy of Sciences (PNAS) last month.
The joint research team’s spatial stochastic model (See Figure 2) described the motion of PER molecules in cells and demonstrated that the PER molecule should be sufficiently condensed around the cell nucleus to be phosphorylated simultaneously and enter the nucleus together (See Figure 3 Left). Thanks to this phosphorylation synchronization switch, thousands of PER molecules can enter the nucleus at the same time every day and maintain stable circadian rhythms.
However, when aging and/or diseases including dementia and obesity cause the cytoplasm to become congested with increased cytoplasmic obstacles such as protein aggregates and fat vacuoles, it hinders the timely condensation of PER molecules around the cell nucleus (See Figure 3 Right). As a result, the phosphorylation synchronization switch does not work and PER proteins enter into the nucleus at irregular times, making the circadian rhythms and sleep-wake cycles unstable, the study revealed.
Professor Kim said, “As a mathematician, I am excited to help enable the advancement of new treatment strategies that can improve the lives of so many patients who suffer from irregular sleep-wake cycles. Taking these findings as an opportunity, I hope to see more active interchanges of ideas and collaboration between mathematical and biological sciences.”
This work was supported by the National Institutes of Health and the National Science Foundation in the US, and the International Human Frontiers Science Program Organization and the National Research Foundation of Korea.
Publication:
Beesley, S. and Kim, D. W, et al. (2020) Wake-sleep cycles are severely disrupted by diseases affecting cytoplasmic homeostasis. Proceedings of the National Academy of Sciences (PNAS), Vol. 117, No. 45, 28402-28411. Available online at https://doi.org/10.1073/pnas.2003524117
Profile:
Jae Kyoung Kim, Ph.D.
Associate Professor
jaekkim@kaist.ac.kr
http://mathsci.kaist.ac.kr/~jaekkim
@umichkim on Twitter
Department of Mathematical Sciences
Korea Advanced Institute of Science and Technology (KAIST)
Daejeon, Republic of Korea
Profile:
Choogon Lee, Ph.D.
Associate Professor
clee@neuro.fsu.edu
https://med.fsu.edu/biosci/lee-lab
Department of Biomedical Sciences
Florida State University
Florida, USA
(END)
2020.12.11 View 11275
Mystery Solved with Math: Cytoplasmic Traffic Jam Disrupts Sleep-Wake Cycles
KAIST mathematicians and their collaborators at Florida State University have identified the principle of how aging and diseases like dementia and obesity cause sleep disorders. A combination of mathematical modelling and experiments demonstrated that the cytoplasmic congestion caused by aging, dementia, and/or obesity disrupts the circadian rhythms in the human body and leads to irregular sleep-wake cycles. This finding suggests new treatment strategies for addressing unstable sleep-wake cycles.
Human bodies adjust sleep schedules in accordance with the ‘circadian rhythms’, which are regulated by our time keeping system, the ‘circadian clock’. This clock tells our body when to rest by generating the 24-hour rhythms of a protein called PERIOD (PER) (See Figure 1).
The amount of the PER protein increases for half of the day and then decreases for the remaining half. The principle is that the PER protein accumulating in the cytoplasm for several hours enters the cell nucleus all at once, hindering the transcription of PER genes and thereby reducing the amount of PER.
However, it has remained a mystery how thousands of PER molecules can simultaneously enter into the nucleus in a complex cell environment where a variety of materials co-exist and can interfere with the motion of PER. This would be like finding a way for thousands of employees from all over New York City to enter an office building at the same time every day.
A group of researchers led by Professor Jae Kyoung Kim from the KAIST Department of Mathematical Sciences solved the mystery by developing a spatiotemporal and probabilistic model that describes the motion of PER molecules in a cell environment.
This study was conducted in collaboration with Professor Choogon Lee’s group from Florida State University, where the experiments were carried out, and the results were published in the Proceedings of the National Academy of Sciences (PNAS) last month.
The joint research team’s spatial stochastic model (See Figure 2) described the motion of PER molecules in cells and demonstrated that the PER molecule should be sufficiently condensed around the cell nucleus to be phosphorylated simultaneously and enter the nucleus together (See Figure 3 Left). Thanks to this phosphorylation synchronization switch, thousands of PER molecules can enter the nucleus at the same time every day and maintain stable circadian rhythms.
However, when aging and/or diseases including dementia and obesity cause the cytoplasm to become congested with increased cytoplasmic obstacles such as protein aggregates and fat vacuoles, it hinders the timely condensation of PER molecules around the cell nucleus (See Figure 3 Right). As a result, the phosphorylation synchronization switch does not work and PER proteins enter into the nucleus at irregular times, making the circadian rhythms and sleep-wake cycles unstable, the study revealed.
Professor Kim said, “As a mathematician, I am excited to help enable the advancement of new treatment strategies that can improve the lives of so many patients who suffer from irregular sleep-wake cycles. Taking these findings as an opportunity, I hope to see more active interchanges of ideas and collaboration between mathematical and biological sciences.”
This work was supported by the National Institutes of Health and the National Science Foundation in the US, and the International Human Frontiers Science Program Organization and the National Research Foundation of Korea.
Publication:
Beesley, S. and Kim, D. W, et al. (2020) Wake-sleep cycles are severely disrupted by diseases affecting cytoplasmic homeostasis. Proceedings of the National Academy of Sciences (PNAS), Vol. 117, No. 45, 28402-28411. Available online at https://doi.org/10.1073/pnas.2003524117
Profile:
Jae Kyoung Kim, Ph.D.
Associate Professor
jaekkim@kaist.ac.kr
http://mathsci.kaist.ac.kr/~jaekkim
@umichkim on Twitter
Department of Mathematical Sciences
Korea Advanced Institute of Science and Technology (KAIST)
Daejeon, Republic of Korea
Profile:
Choogon Lee, Ph.D.
Associate Professor
clee@neuro.fsu.edu
https://med.fsu.edu/biosci/lee-lab
Department of Biomedical Sciences
Florida State University
Florida, USA
(END)
2020.12.11 View 11275 -
 Drawing the Line to Answer Art’s Big Questions
- KAIST scientists show how statistical physics can reveal art trends across time and culture. -
Algorithms have shown that the compositional structure of Western landscape paintings changed “suspiciously” smoothly between 1500 and 2000 AD, potentially indicating a selection bias by art curators or in art historical literature, physicists from the Korea Advanced Institute of Science and Technology (KAIST) and colleagues report in the Proceedings of the National Academy of Sciences (PNAS).
KAIST statistical physicist Hawoong Jeong worked with statisticians, digital analysts and art historians in Korea, Estonia and the US to clarify whether computer algorithms could help resolve long-standing questions about design principles used in landscape paintings, such as the placement of the horizon and other primary features.
“A foundational question among art historians is whether artwork contains organizing principles that transcend culture and time and, if yes, how these principles evolved over time,” explains Jeong. “We developed an information-theoretic approach that can capture compositional proportion in landscape paintings and found that the preferred compositional proportion systematically evolved over time.”
Digital versions of almost 15,000 canonical landscape paintings from the Western renaissance in the 1500s to the more recent contemporary art period were run through a computer algorithm. The algorithm progressively divides artwork into horizontal and vertical lines depending on the amount of information in each subsequent partition. It allows scientists to evaluate how artists and various art styles compose landscape artwork, in terms of placement of a piece’s most important components, in addition to how high or low the landscape’s horizon is placed.
The scientists started by analysing the first two partitioning lines identified by the algorithm in the paintings and found they could be categorized into four groups: an initial horizontal line followed by a second horizontal line (H-H); an initial horizontal line followed by a second vertical line (H-V); a vertical followed by horizontal line (V-H); or a vertical followed by a vertical line (V-V) (see image 1 and 2). They then looked at the categorizations over time.
They found that before the mid-nineteenth century, H-V was the dominant composition type, followed by H-H, V-H, and V-V. The mid-nineteenth century then brought change, with the H-V composition style decreasing in popularity with a rise in the H-H composition style. The other two styles remained relatively stable.
The scientists also looked at how the horizon line, which separates sky from land, changed over time. In the 16th century, the dominant horizon line of the painting was above the middle of the canvas, but it gradually descended to the lower middle of the canvas by the 17th century, where it remained until the mid-nineteenth century. After that, the horizon line began gradually rising again.
Interestingly, the algorithm showed that these findings were similar across cultures and artistic periods, even through periods dominated by a diversity in art styles. This similarity may well be a function, then, of a bias in the dataset.
“In recent decades, art historians have prioritized the argument that there is great diversity in the evolution of artistic expression rather than offering a relatively smoother consensus story in Western art,” Jeong says. “This study serves as a reminder that the available large-scale datasets might be perpetuating severe biases.”
The scientists next aim to broaden their analyses to include more diverse artwork, as this particular dataset was ultimately Western and male biased. Future analyses should also consider diagonal compositions in paintings, they say.
This work was supported by the National Research Foundation (NRF) of Korea.
Publication:
Lee, B, et al. (2020) Dissecting landscape art history with information theory. Proceedings of the National Academy of Sciences (PNAS), Vol. 117, No. 43, 26580-26590. Available online at https://doi.org/10.1073/pnas.2011927117
Profile:
Hawoong Jeong, Ph.D.
Professor
hjeong@kaist.ac.kr
https://www.kaist.ac.kr
Department of Physics
Korea Advanced Institute of Science and Technology (KAIST)
Daejeon, Republic of Korea
(END)
2020.11.13 View 12214
Drawing the Line to Answer Art’s Big Questions
- KAIST scientists show how statistical physics can reveal art trends across time and culture. -
Algorithms have shown that the compositional structure of Western landscape paintings changed “suspiciously” smoothly between 1500 and 2000 AD, potentially indicating a selection bias by art curators or in art historical literature, physicists from the Korea Advanced Institute of Science and Technology (KAIST) and colleagues report in the Proceedings of the National Academy of Sciences (PNAS).
KAIST statistical physicist Hawoong Jeong worked with statisticians, digital analysts and art historians in Korea, Estonia and the US to clarify whether computer algorithms could help resolve long-standing questions about design principles used in landscape paintings, such as the placement of the horizon and other primary features.
“A foundational question among art historians is whether artwork contains organizing principles that transcend culture and time and, if yes, how these principles evolved over time,” explains Jeong. “We developed an information-theoretic approach that can capture compositional proportion in landscape paintings and found that the preferred compositional proportion systematically evolved over time.”
Digital versions of almost 15,000 canonical landscape paintings from the Western renaissance in the 1500s to the more recent contemporary art period were run through a computer algorithm. The algorithm progressively divides artwork into horizontal and vertical lines depending on the amount of information in each subsequent partition. It allows scientists to evaluate how artists and various art styles compose landscape artwork, in terms of placement of a piece’s most important components, in addition to how high or low the landscape’s horizon is placed.
The scientists started by analysing the first two partitioning lines identified by the algorithm in the paintings and found they could be categorized into four groups: an initial horizontal line followed by a second horizontal line (H-H); an initial horizontal line followed by a second vertical line (H-V); a vertical followed by horizontal line (V-H); or a vertical followed by a vertical line (V-V) (see image 1 and 2). They then looked at the categorizations over time.
They found that before the mid-nineteenth century, H-V was the dominant composition type, followed by H-H, V-H, and V-V. The mid-nineteenth century then brought change, with the H-V composition style decreasing in popularity with a rise in the H-H composition style. The other two styles remained relatively stable.
The scientists also looked at how the horizon line, which separates sky from land, changed over time. In the 16th century, the dominant horizon line of the painting was above the middle of the canvas, but it gradually descended to the lower middle of the canvas by the 17th century, where it remained until the mid-nineteenth century. After that, the horizon line began gradually rising again.
Interestingly, the algorithm showed that these findings were similar across cultures and artistic periods, even through periods dominated by a diversity in art styles. This similarity may well be a function, then, of a bias in the dataset.
“In recent decades, art historians have prioritized the argument that there is great diversity in the evolution of artistic expression rather than offering a relatively smoother consensus story in Western art,” Jeong says. “This study serves as a reminder that the available large-scale datasets might be perpetuating severe biases.”
The scientists next aim to broaden their analyses to include more diverse artwork, as this particular dataset was ultimately Western and male biased. Future analyses should also consider diagonal compositions in paintings, they say.
This work was supported by the National Research Foundation (NRF) of Korea.
Publication:
Lee, B, et al. (2020) Dissecting landscape art history with information theory. Proceedings of the National Academy of Sciences (PNAS), Vol. 117, No. 43, 26580-26590. Available online at https://doi.org/10.1073/pnas.2011927117
Profile:
Hawoong Jeong, Ph.D.
Professor
hjeong@kaist.ac.kr
https://www.kaist.ac.kr
Department of Physics
Korea Advanced Institute of Science and Technology (KAIST)
Daejeon, Republic of Korea
(END)
2020.11.13 View 12214 -
 X-ray Scattering Shines Light on Protein Folding
- Multiple forms of a non-functional, unfolded protein follow different pathways and timelines to reach its folded, functional state, a study reveals. -
KAIST researchers have used an X-ray method to track how proteins fold, which could improve computer simulations of this process, with implications for understanding diseases and improving drug discovery. Their findings were reported in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 30.
When proteins are translated from their DNA codes, they quickly transform from a non-functional, unfolded state into their folded, functional state. Problems in folding can lead to diseases like Alzheimer’s and Parkinson’s.
“Protein folding is one of the most important biological processes, as it forms the functioning 3D protein structure,” explained the physical chemist Hyotcherl Ihee of the Department of Chemistry at KAIST. Dr. Tae Wu Kim, the lead author of this research from Ihee’s group, added, “Understanding the mechanisms of protein folding is important, and could pave the way for disease study and drug development.”
Ihee’s team developed an approach using an X-ray scattering technique to uncover how the protein cytochrome c folds from its initial unfolded state. This protein is composed of a chain of 104 amino acids with an iron-containing heme molecule. It is often used for protein folding studies.
The researchers placed the protein in a solution and shined ultraviolet light on it. This process provides electrons to cytochrome c, reducing the iron within it from the ferric to the ferrous form, which initiates folding. As this was happening, the researchers beamed X-rays at very short intervals onto the sample. The X-rays scattered off all the atomic pairs in the sample and a detector continuously recorded the X-ray scattering patterns. The X-ray scattering patterns provided direct information regarding the 3D protein structure and the changes made in these patterns over time showed real-time motion of the protein during the folding process.
The team found cytochrome c proteins initially exist in a wide variety of unfolded states. Once the folding process is triggered, they stop by a group of intermediates within 31.6 microseconds, and then those intermediates follow different pathways with different folding times to reach an energetically stable folded state.
“We don’t know if this diversity in folding paths can be generalized to other proteins,” Ihee confessed. He continued, “However, we believe that our approach can be used to study other protein folding systems.”
Ihee hopes this approach can improve the accuracy of models that simulate protein interactions by including information on their unstructured states. These simulations are important as they can help identify barriers to proper folding and predict a protein’s folded state given its amino acid sequence. Ultimately, the models could help clarify how some diseases develop and how drugs interact with various protein structures.
Ihee’s group collaborated with Professor Young Min Rhee at the KAIST Department of Chemistry, and this work was supported by the National Research Foundation of Korea (NRF) and the Institute for Basic Science (IBS).
Figure. The scientists found that non-functional unfolded forms of the protein cytochrome c follow different pathways and timelines to reach a stable functional folded state.
Publications:
Kim, T. W., et al. (2020) ‘Protein folding from heterogeneous unfolded state revealed by time-resolved X-ray solution scattering’. PNAS. Volume 117. Issue 26. Page 14996-15005. Available online at https://doi.org/10.1073/pnas.1913442117
Profile: Hyotcherl Ihee, Ph.D.
Professor
hyotcherl.ihee@kaist.ac.kr
http://time.kaist.ac.kr/
Ihee Laboratory
Department of Chemistry
KAIST
https://www.kaist.ac.kr
Daejeon 34141, Korea
Profile: Young Min Rhee, Ph.D.
Professor
ymrhee@kaist.ac.kr
http://singlet.kaist.ac.kr
Rhee Research Group
Department of Chemistry
KAIST
https://www.kaist.ac.kr
Daejeon 34141, Korea
(END)
2020.07.09 View 16168
X-ray Scattering Shines Light on Protein Folding
- Multiple forms of a non-functional, unfolded protein follow different pathways and timelines to reach its folded, functional state, a study reveals. -
KAIST researchers have used an X-ray method to track how proteins fold, which could improve computer simulations of this process, with implications for understanding diseases and improving drug discovery. Their findings were reported in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 30.
When proteins are translated from their DNA codes, they quickly transform from a non-functional, unfolded state into their folded, functional state. Problems in folding can lead to diseases like Alzheimer’s and Parkinson’s.
“Protein folding is one of the most important biological processes, as it forms the functioning 3D protein structure,” explained the physical chemist Hyotcherl Ihee of the Department of Chemistry at KAIST. Dr. Tae Wu Kim, the lead author of this research from Ihee’s group, added, “Understanding the mechanisms of protein folding is important, and could pave the way for disease study and drug development.”
Ihee’s team developed an approach using an X-ray scattering technique to uncover how the protein cytochrome c folds from its initial unfolded state. This protein is composed of a chain of 104 amino acids with an iron-containing heme molecule. It is often used for protein folding studies.
The researchers placed the protein in a solution and shined ultraviolet light on it. This process provides electrons to cytochrome c, reducing the iron within it from the ferric to the ferrous form, which initiates folding. As this was happening, the researchers beamed X-rays at very short intervals onto the sample. The X-rays scattered off all the atomic pairs in the sample and a detector continuously recorded the X-ray scattering patterns. The X-ray scattering patterns provided direct information regarding the 3D protein structure and the changes made in these patterns over time showed real-time motion of the protein during the folding process.
The team found cytochrome c proteins initially exist in a wide variety of unfolded states. Once the folding process is triggered, they stop by a group of intermediates within 31.6 microseconds, and then those intermediates follow different pathways with different folding times to reach an energetically stable folded state.
“We don’t know if this diversity in folding paths can be generalized to other proteins,” Ihee confessed. He continued, “However, we believe that our approach can be used to study other protein folding systems.”
Ihee hopes this approach can improve the accuracy of models that simulate protein interactions by including information on their unstructured states. These simulations are important as they can help identify barriers to proper folding and predict a protein’s folded state given its amino acid sequence. Ultimately, the models could help clarify how some diseases develop and how drugs interact with various protein structures.
Ihee’s group collaborated with Professor Young Min Rhee at the KAIST Department of Chemistry, and this work was supported by the National Research Foundation of Korea (NRF) and the Institute for Basic Science (IBS).
Figure. The scientists found that non-functional unfolded forms of the protein cytochrome c follow different pathways and timelines to reach a stable functional folded state.
Publications:
Kim, T. W., et al. (2020) ‘Protein folding from heterogeneous unfolded state revealed by time-resolved X-ray solution scattering’. PNAS. Volume 117. Issue 26. Page 14996-15005. Available online at https://doi.org/10.1073/pnas.1913442117
Profile: Hyotcherl Ihee, Ph.D.
Professor
hyotcherl.ihee@kaist.ac.kr
http://time.kaist.ac.kr/
Ihee Laboratory
Department of Chemistry
KAIST
https://www.kaist.ac.kr
Daejeon 34141, Korea
Profile: Young Min Rhee, Ph.D.
Professor
ymrhee@kaist.ac.kr
http://singlet.kaist.ac.kr
Rhee Research Group
Department of Chemistry
KAIST
https://www.kaist.ac.kr
Daejeon 34141, Korea
(END)
2020.07.09 View 16168 -
 3D Hierarchically Porous Nanostructured Catalyst Helps Efficiently Reduce CO2
- This new catalyst will bring CO2 one step closer to serving as a sustainable energy source. -
KAIST researchers developed a three-dimensional (3D) hierarchically porous nanostructured catalyst with carbon dioxide (CO2) to carbon monoxide (CO) conversion rate up to 3.96 times higher than that of conventional nanoporous gold catalysts. This new catalyst helps overcome the existing limitations of the mass transport that has been a major cause of decreases in the CO2 conversion rate, holding a strong promise for the large-scale and cost-effective electrochemical conversion of CO2 into useful chemicals.
As CO2 emissions increase and fossil fuels deplete globally, reducing and converting CO2 to clean energy electrochemically has attracted a great deal of attention as a promising technology. Especially due to the fact that the CO2 reduction reaction occurs competitively with hydrogen evolution reactions (HER) at similar redox potentials, the development of an efficient electrocatalyst for selective and robust CO2 reduction reactions has remained a key technological issue.
Gold (Au) is one of the most commonly used catalysts in CO2 reduction reactions, but the high cost and scarcity of Au pose obstacles for mass commercial applications. The development of nanostructures has been extensively studied as a potential approach to improving the selectivity for target products and maximizing the number of active stable sites, thus enhancing the energy efficiency.
However, the nanopores of the previously reported complex nanostructures were easily blocked by gaseous CO bubbles during aqueous reactions. The CO bubbles hindered mass transport of the reactants through the electrolyte, resulting in low CO2 conversion rates.
In the study published in the Proceedings of the National Academy of Sciences of the USA (PNAS) on March 4, a research group at KAIST led by Professor Seokwoo Jeon and Professor Jihun Oh from the Department of Materials Science and Engineering designed a 3D hierarchically porous Au nanostructure with two different sizes of macropores and nanopores. The team used proximity-field nanopatterning (PnP) and electroplating techniques that are effective for fabricating the 3D well-ordered nanostructures.
The proposed nanostructure, comprised of interconnected macroporous channels 200 to 300 nanometers (nm) wide and 10 nm nanopores, induces efficient mass transport through the interconnected macroporous channels as well as high selectivity by producing highly active stable sites from numerous nanopores.
As a result, its electrodes show a high CO selectivity of 85.8% at a low overpotential of 0.264 V and efficient mass activity that is up to 3.96 times higher than that of de-alloyed nanoporous Au electrodes.
“These results are expected to solve the problem of mass transfer in the field of similar electrochemical reactions and can be applied to a wide range of green energy applications for the efficient utilization of electrocatalysts,” said the researchers.
This work was supported by the National Research Foundation (NRF) of Korea.
Image credit: Professor Seokwoo Jeon and Professor Jihun Oh, KAIST
Image usage restrictions: News organizations may use or redistribute this image, with proper attribution, as part of news coverage of this paper only.
Publication:
Hyun et al. (2020) Hierarchically porous Au nanostructures with interconnected channels for efficient mass transport in electrocatalytic CO2 reduction. Proceedings of the National Academy of Sciences of the USA (PNAS). Available online at https://doi.org/10.1073/pnas.1918837117
Profile:
Seokwoo Jeon, PhD
Professor
jeon39@kaist.ac.kr
http://fdml.kaist.ac.kr
Department of Materials Science and Engineering (MSE)
https://www.kaist.ac.kr
Korea Advanced Institute of Science and Technology (KAIST)Daejeon, Republic of Korea
Profile:
Jihun Oh, PhD
Associate Professor
jihun.oh@kaist.ac.kr
http://les.kaist.ac.kr
Department of Materials Science and Engineering (MSE)
Department of Energy, Environment, Water and Sustainability (EEWS)
KAIST
Profile:
Gayea Hyun
PhD Candidate
cldywkd93@kaist.ac.kr
http://fdml.kaist.ac.kr
Flexible Devices and Metamaterials Laboratory (FDML)
Department of Materials Science and Engineering (MSE)
KAIST
Profile:
Jun Tae Song, PhD
Assistant Professor
song.juntae@cstf.kyushu-u.ac.jp
http://www.cstf.kyushu-u.ac.jp/~ishihara-lab/
Department of Applied Chemistry
https://www.kyushu-u.ac.jp
Kyushu UniversityFukuoka, Japan
(END)
2020.03.13 View 18642
3D Hierarchically Porous Nanostructured Catalyst Helps Efficiently Reduce CO2
- This new catalyst will bring CO2 one step closer to serving as a sustainable energy source. -
KAIST researchers developed a three-dimensional (3D) hierarchically porous nanostructured catalyst with carbon dioxide (CO2) to carbon monoxide (CO) conversion rate up to 3.96 times higher than that of conventional nanoporous gold catalysts. This new catalyst helps overcome the existing limitations of the mass transport that has been a major cause of decreases in the CO2 conversion rate, holding a strong promise for the large-scale and cost-effective electrochemical conversion of CO2 into useful chemicals.
As CO2 emissions increase and fossil fuels deplete globally, reducing and converting CO2 to clean energy electrochemically has attracted a great deal of attention as a promising technology. Especially due to the fact that the CO2 reduction reaction occurs competitively with hydrogen evolution reactions (HER) at similar redox potentials, the development of an efficient electrocatalyst for selective and robust CO2 reduction reactions has remained a key technological issue.
Gold (Au) is one of the most commonly used catalysts in CO2 reduction reactions, but the high cost and scarcity of Au pose obstacles for mass commercial applications. The development of nanostructures has been extensively studied as a potential approach to improving the selectivity for target products and maximizing the number of active stable sites, thus enhancing the energy efficiency.
However, the nanopores of the previously reported complex nanostructures were easily blocked by gaseous CO bubbles during aqueous reactions. The CO bubbles hindered mass transport of the reactants through the electrolyte, resulting in low CO2 conversion rates.
In the study published in the Proceedings of the National Academy of Sciences of the USA (PNAS) on March 4, a research group at KAIST led by Professor Seokwoo Jeon and Professor Jihun Oh from the Department of Materials Science and Engineering designed a 3D hierarchically porous Au nanostructure with two different sizes of macropores and nanopores. The team used proximity-field nanopatterning (PnP) and electroplating techniques that are effective for fabricating the 3D well-ordered nanostructures.
The proposed nanostructure, comprised of interconnected macroporous channels 200 to 300 nanometers (nm) wide and 10 nm nanopores, induces efficient mass transport through the interconnected macroporous channels as well as high selectivity by producing highly active stable sites from numerous nanopores.
As a result, its electrodes show a high CO selectivity of 85.8% at a low overpotential of 0.264 V and efficient mass activity that is up to 3.96 times higher than that of de-alloyed nanoporous Au electrodes.
“These results are expected to solve the problem of mass transfer in the field of similar electrochemical reactions and can be applied to a wide range of green energy applications for the efficient utilization of electrocatalysts,” said the researchers.
This work was supported by the National Research Foundation (NRF) of Korea.
Image credit: Professor Seokwoo Jeon and Professor Jihun Oh, KAIST
Image usage restrictions: News organizations may use or redistribute this image, with proper attribution, as part of news coverage of this paper only.
Publication:
Hyun et al. (2020) Hierarchically porous Au nanostructures with interconnected channels for efficient mass transport in electrocatalytic CO2 reduction. Proceedings of the National Academy of Sciences of the USA (PNAS). Available online at https://doi.org/10.1073/pnas.1918837117
Profile:
Seokwoo Jeon, PhD
Professor
jeon39@kaist.ac.kr
http://fdml.kaist.ac.kr
Department of Materials Science and Engineering (MSE)
https://www.kaist.ac.kr
Korea Advanced Institute of Science and Technology (KAIST)Daejeon, Republic of Korea
Profile:
Jihun Oh, PhD
Associate Professor
jihun.oh@kaist.ac.kr
http://les.kaist.ac.kr
Department of Materials Science and Engineering (MSE)
Department of Energy, Environment, Water and Sustainability (EEWS)
KAIST
Profile:
Gayea Hyun
PhD Candidate
cldywkd93@kaist.ac.kr
http://fdml.kaist.ac.kr
Flexible Devices and Metamaterials Laboratory (FDML)
Department of Materials Science and Engineering (MSE)
KAIST
Profile:
Jun Tae Song, PhD
Assistant Professor
song.juntae@cstf.kyushu-u.ac.jp
http://www.cstf.kyushu-u.ac.jp/~ishihara-lab/
Department of Applied Chemistry
https://www.kyushu-u.ac.jp
Kyushu UniversityFukuoka, Japan
(END)
2020.03.13 View 18642 -
 Recombinant E. Coli As a Biofactory for the Biosynthesis of Diverse Nanomaterials
(Distinguished Professor Lee and PhD candidate Choi)
A metabolic research group at KAIST and Chung-Ang University in Korea has developed a recombinant E. coli strain that biosynthesizes 60 different nanomaterials covering 35 elements on the periodic table. Among the elements, the team could biosynthesize 33 novel nanomaterials for the first time, advancing the forward design of nanomaterials through the biosynthesis of various single and multi-elements.
The study analyzed the nanomaterial biosynthesis conditions using a Pourbaix diagram to predict the producibility and crystallinity. Researchers studied a Pourbaix diagram to predict the stable chemical species of each element for nanomaterial biosynthesis at varying levels of reduction potential (Eh) and pH. Based on the Pourbaix diagram analyses, the initial pH of the reaction was changed from 6.5 to 7.5, resulting in the biosynthesis of various crystalline nanomaterials that were previously amorphous or not synthesized.
This strategy was extended to biosynthesize multi-element nanomaterials. Various single and multi-element nanomaterials biosynthesized in this research can potentially serve as new and novel nanomaterials for industrial applications such as catalysts, chemical sensors, biosensors, bioimaging, drug delivery, and cancer therapy.
A research group consisting of PhD candidate Yoojin Choi, Associate Professor Doh Chang Lee, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST and Associate Professor Tae Jung Park of the Department of Chemistry at Chung-Ang University reported the synthesis. This study, entitled “Recombinant Escherichia coli as a biofactory for various single- and multi-element nanomaterials,” was published online in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on May 21.
A recent successful biosynthesis of nanomaterials under mild conditions without requiring physical and chemical treatments has triggered the exploration of the full biosynthesis capacity of a biological system for producing a diverse range of nanomaterials as well as for understanding biosynthesis mechanisms for crystalline versus amorphous nanomaterials.
There has been increased interest in synthesizing various nanomaterials that have not yet been synthesized for various applications including semiconducting materials, enhanced solar cells, biomedical materials, and many others. This research reports the construction of a recombinant E. coli strain that co-expresses metallothionein, a metal binding protein, and phytochelatin synthase that synthesizes the metal-binding peptide phytochelatin for the biosynthesis of various nanomaterials. Subsequently, an E. coli strain was engineered to produce a diverse range of nanomaterials, including those never biosynthesized before, by using 35 individual elements from the periodic table and also by combining multi-elements.
Distinguished Professor Lee said, “An environmentally-friendly and sustainable process is of much interest for producing nanomaterials by not only chemical and physical methods but biological synthesis. Moreover, there has been much attention paid to producing diverse and novel nanomaterials for new industrial applications. This is the first report to predict the biosynthesis of various nanomaterials, by far the largest number of various single- and multi-elements nanomaterials. The strategies used for nanomaterial biosynthesis in this research will be useful for further diversifying the portfolio of nanomaterials that can be manufactured.”
Figure: The biosynthesis of diverse nanomaterials using recombinant E. coli. This schematic diagram shows the overall conceptualization of the biosynthesis of various single and multi-element nanomaterials using recombinant E. coli under incubation with corresponding elemental precursors. The 35 elements that were tested to biosynthesize nanomaterials are shown in black circles on the periodic table.
2018.05.23 View 12110
Recombinant E. Coli As a Biofactory for the Biosynthesis of Diverse Nanomaterials
(Distinguished Professor Lee and PhD candidate Choi)
A metabolic research group at KAIST and Chung-Ang University in Korea has developed a recombinant E. coli strain that biosynthesizes 60 different nanomaterials covering 35 elements on the periodic table. Among the elements, the team could biosynthesize 33 novel nanomaterials for the first time, advancing the forward design of nanomaterials through the biosynthesis of various single and multi-elements.
The study analyzed the nanomaterial biosynthesis conditions using a Pourbaix diagram to predict the producibility and crystallinity. Researchers studied a Pourbaix diagram to predict the stable chemical species of each element for nanomaterial biosynthesis at varying levels of reduction potential (Eh) and pH. Based on the Pourbaix diagram analyses, the initial pH of the reaction was changed from 6.5 to 7.5, resulting in the biosynthesis of various crystalline nanomaterials that were previously amorphous or not synthesized.
This strategy was extended to biosynthesize multi-element nanomaterials. Various single and multi-element nanomaterials biosynthesized in this research can potentially serve as new and novel nanomaterials for industrial applications such as catalysts, chemical sensors, biosensors, bioimaging, drug delivery, and cancer therapy.
A research group consisting of PhD candidate Yoojin Choi, Associate Professor Doh Chang Lee, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST and Associate Professor Tae Jung Park of the Department of Chemistry at Chung-Ang University reported the synthesis. This study, entitled “Recombinant Escherichia coli as a biofactory for various single- and multi-element nanomaterials,” was published online in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on May 21.
A recent successful biosynthesis of nanomaterials under mild conditions without requiring physical and chemical treatments has triggered the exploration of the full biosynthesis capacity of a biological system for producing a diverse range of nanomaterials as well as for understanding biosynthesis mechanisms for crystalline versus amorphous nanomaterials.
There has been increased interest in synthesizing various nanomaterials that have not yet been synthesized for various applications including semiconducting materials, enhanced solar cells, biomedical materials, and many others. This research reports the construction of a recombinant E. coli strain that co-expresses metallothionein, a metal binding protein, and phytochelatin synthase that synthesizes the metal-binding peptide phytochelatin for the biosynthesis of various nanomaterials. Subsequently, an E. coli strain was engineered to produce a diverse range of nanomaterials, including those never biosynthesized before, by using 35 individual elements from the periodic table and also by combining multi-elements.
Distinguished Professor Lee said, “An environmentally-friendly and sustainable process is of much interest for producing nanomaterials by not only chemical and physical methods but biological synthesis. Moreover, there has been much attention paid to producing diverse and novel nanomaterials for new industrial applications. This is the first report to predict the biosynthesis of various nanomaterials, by far the largest number of various single- and multi-elements nanomaterials. The strategies used for nanomaterial biosynthesis in this research will be useful for further diversifying the portfolio of nanomaterials that can be manufactured.”
Figure: The biosynthesis of diverse nanomaterials using recombinant E. coli. This schematic diagram shows the overall conceptualization of the biosynthesis of various single and multi-element nanomaterials using recombinant E. coli under incubation with corresponding elemental precursors. The 35 elements that were tested to biosynthesize nanomaterials are shown in black circles on the periodic table.
2018.05.23 View 12110 -
 Professor Seyun Kim Identifies a Neuron Signal Controlling Molecule
A research team led by Professor Seyun Kim of the Department of Biological Sciences at KAIST has identified inositol pyrophosphates as the molecule that strongly controls neuron signaling via synaptotagmin.
Professors Tae-Young Yoon of Yonsei University’s Y-IBS and Sung-Hyun Kim of Kyung Hee University’s Department of Biomedical Science also joined the team.
The results were published in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 30, 2016.
This interdisciplinary research project was conducted by six research teams from four different countries and covered a wide scope of academic fields, from neurobiology to super resolution optic imaging.
Inositol pyrophosphates such as 5-diphosphoinositol pentakisphos-phate (5-IP7), which naturally occur in corns and beans, are essential metabolites in the body. In particular, inositol hexakisphosphate (IP6) has anti-cancer properties and is thought to have an important role in cell signaling.
Inositol pentakisphosphate (IP7) differs from IP6 by having an additional phosphate group, which was first discovered 20 years ago. IP7 has recently been identified as playing a key role in diabetes and obesity.
Psychopathy and neurodegenerative diseases are known to result from the disrupted balance of inositol pyrophosphates. However, the role and the mechanism of action of IP7 in brain neurons and nerve transmission remained unknown.
Professor Kim’s team has worked on inositol pyrophosphates for several years and discovered that very small quantities of IP7 control cell-signaling transduction. Professor Yoon of Yonsei University identified IP7 as a much stronger inhibitor of neuron signaling compared to IP6. In particular, IP7 directly suppresses synaptotagmin, one of the key proteins in neuron signaling. Moreover, Professor Kim of Kyung Hee University observed IP7 inhibition in sea horse neurons.
Together, the joint research team identified inositol pyrophosphates as the key switch metabolite of brain-signaling transduction.
The researchers hope that future research on synaptotagmin and IP7 will reveal the mechanism of neuron-signal transduction and thus enable the treatment of neurological disorders.
These research findings were the result of cooperation of various science and technology institutes: KAIST, Yonsei-IBS (Institute for Basic Science), Kyung Hee University, Sungkyunkwan University, KIST, University of Zurich in Switzerland, and Albert-Ludwigs-University Freiburg in Germany.
Schematic Image of Controlling the Synaptic Exocytotic Pathway by 5-IP7 , Helping the Understanding of the Signaling Mechanisms of Inositol Pyrophosphates
2016.07.21 View 12518
Professor Seyun Kim Identifies a Neuron Signal Controlling Molecule
A research team led by Professor Seyun Kim of the Department of Biological Sciences at KAIST has identified inositol pyrophosphates as the molecule that strongly controls neuron signaling via synaptotagmin.
Professors Tae-Young Yoon of Yonsei University’s Y-IBS and Sung-Hyun Kim of Kyung Hee University’s Department of Biomedical Science also joined the team.
The results were published in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 30, 2016.
This interdisciplinary research project was conducted by six research teams from four different countries and covered a wide scope of academic fields, from neurobiology to super resolution optic imaging.
Inositol pyrophosphates such as 5-diphosphoinositol pentakisphos-phate (5-IP7), which naturally occur in corns and beans, are essential metabolites in the body. In particular, inositol hexakisphosphate (IP6) has anti-cancer properties and is thought to have an important role in cell signaling.
Inositol pentakisphosphate (IP7) differs from IP6 by having an additional phosphate group, which was first discovered 20 years ago. IP7 has recently been identified as playing a key role in diabetes and obesity.
Psychopathy and neurodegenerative diseases are known to result from the disrupted balance of inositol pyrophosphates. However, the role and the mechanism of action of IP7 in brain neurons and nerve transmission remained unknown.
Professor Kim’s team has worked on inositol pyrophosphates for several years and discovered that very small quantities of IP7 control cell-signaling transduction. Professor Yoon of Yonsei University identified IP7 as a much stronger inhibitor of neuron signaling compared to IP6. In particular, IP7 directly suppresses synaptotagmin, one of the key proteins in neuron signaling. Moreover, Professor Kim of Kyung Hee University observed IP7 inhibition in sea horse neurons.
Together, the joint research team identified inositol pyrophosphates as the key switch metabolite of brain-signaling transduction.
The researchers hope that future research on synaptotagmin and IP7 will reveal the mechanism of neuron-signal transduction and thus enable the treatment of neurological disorders.
These research findings were the result of cooperation of various science and technology institutes: KAIST, Yonsei-IBS (Institute for Basic Science), Kyung Hee University, Sungkyunkwan University, KIST, University of Zurich in Switzerland, and Albert-Ludwigs-University Freiburg in Germany.
Schematic Image of Controlling the Synaptic Exocytotic Pathway by 5-IP7 , Helping the Understanding of the Signaling Mechanisms of Inositol Pyrophosphates
2016.07.21 View 12518 -
 Artificial Antibody-based Therapeutic Candidate for Lung Cancer Developed
Professor Hak-Sung Kim of Biological Sciences at KAIST publishes a cover article on artificial antibody in "Molecular Therapy".
Repebody-based lung cancer therapeutic drug candidate developed Repebody-based protein demonstrates the possibility of the development of a new drug
KAIST Biological Sciences Department’s Professor Hak-Sung Kim, in collaboration with Professor Eun-Kyung Cho from the College of Medicine at Chungnam National University, has successfully developed an artificial antibody-based, or repebody, cancer therapeutic candidate. These research results were published as a cover paper of the July edition of Molecular Therapy.
The repebody developed by Professor Kim and his team strongly binds to interleukin-6, a cancer-causing factor. It has also been confirmed that the repebody can significantly inhibit the proliferation of cancer cells in non-small-cell lung cancer animal model.
Numerous multinational pharmaceutical and biotechnology companies have invested astronomical amounts of money in research for the development of protein therapeutics with low side effects and high efficacy. More than 20 kinds of such therapeutics are currently under clinical trials, and over 100 drugs are under clinical demonstration. Among these, the majority is antibody-based therapeutics, and most of the investments are heavily concentrated in this field. However, antibody production cost is very high because it has large molecular weights and complex structural properties, and this makes it difficult to engineer. Consequently, the development costs a great deal of time and money.
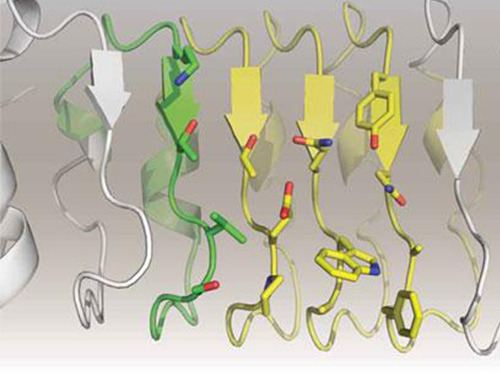
In order to overcome the existing limitations of antibody-based therapeutics, Professor Kim and his team have developed a new artificial antibody, or repebody, which was published in Proceedings of the National Academy of Sciences (PNAS) in 2012. Based on this research, they have succeeded in developing a therapeutic candidate for treating non-small-cell lung cancer with a specifically strong cohesion to the cancer-causing factor, interleukin-6.
Interleukin-6 is a crucial substance within the body that is involved in immune and inflammatory-related signals. When abnormally expressed, it activates various carcinogenic pathways and promotes tumor growth and metastasis. Because of its importance, multinational pharmaceutical companies are heavily investing in developing therapeutics that can inhibit the signaling of interleukin-6.
In this study, Professor Kim and his team observed that a repebody consists of repeated modules, and they conceived a module-based affinity amplification technology that can effectively increase the binding affinity with the disease target. The developed therapeutic candidate has been confirmed in cell and animal experiments to show low immunogenicity, as well as to strongly inhibit the proliferation of non-small-cell lung cancer.
Furthermore, by investigating the complex structure of the repebody with interleukin-6, Professor Kim has identified its mechanism, which demonstrated the potential for therapeutic development. The researchers are currently carrying out pre-clinical trials for acquiring permission to perform clinical trials on animals with non-small-cell lung cancer. The repebody can be developed into a new protein drug after demonstrating its safety and efficacy.
Professor Hak-Sung Kim and his team have confirmed that the repebody can be utilized as a new protein drug, and this will be a significant contribution to Korea’s protein drugs and biotechnology industry development.
The research was supported by the Future Pioneer Industry project and sponsored by the Ministry of Science, ICT and Future Planning.
Figure 1. Professor Kim’s article published as the cover article of July edition of Molecular Therapy
Figure 2. Clinical proof of the repebody’s inhibition of cancer growth using animal models
2014.07.14 View 13992
Artificial Antibody-based Therapeutic Candidate for Lung Cancer Developed
Professor Hak-Sung Kim of Biological Sciences at KAIST publishes a cover article on artificial antibody in "Molecular Therapy".
Repebody-based lung cancer therapeutic drug candidate developed Repebody-based protein demonstrates the possibility of the development of a new drug
KAIST Biological Sciences Department’s Professor Hak-Sung Kim, in collaboration with Professor Eun-Kyung Cho from the College of Medicine at Chungnam National University, has successfully developed an artificial antibody-based, or repebody, cancer therapeutic candidate. These research results were published as a cover paper of the July edition of Molecular Therapy.
The repebody developed by Professor Kim and his team strongly binds to interleukin-6, a cancer-causing factor. It has also been confirmed that the repebody can significantly inhibit the proliferation of cancer cells in non-small-cell lung cancer animal model.
Numerous multinational pharmaceutical and biotechnology companies have invested astronomical amounts of money in research for the development of protein therapeutics with low side effects and high efficacy. More than 20 kinds of such therapeutics are currently under clinical trials, and over 100 drugs are under clinical demonstration. Among these, the majority is antibody-based therapeutics, and most of the investments are heavily concentrated in this field. However, antibody production cost is very high because it has large molecular weights and complex structural properties, and this makes it difficult to engineer. Consequently, the development costs a great deal of time and money.
In order to overcome the existing limitations of antibody-based therapeutics, Professor Kim and his team have developed a new artificial antibody, or repebody, which was published in Proceedings of the National Academy of Sciences (PNAS) in 2012. Based on this research, they have succeeded in developing a therapeutic candidate for treating non-small-cell lung cancer with a specifically strong cohesion to the cancer-causing factor, interleukin-6.
Interleukin-6 is a crucial substance within the body that is involved in immune and inflammatory-related signals. When abnormally expressed, it activates various carcinogenic pathways and promotes tumor growth and metastasis. Because of its importance, multinational pharmaceutical companies are heavily investing in developing therapeutics that can inhibit the signaling of interleukin-6.
In this study, Professor Kim and his team observed that a repebody consists of repeated modules, and they conceived a module-based affinity amplification technology that can effectively increase the binding affinity with the disease target. The developed therapeutic candidate has been confirmed in cell and animal experiments to show low immunogenicity, as well as to strongly inhibit the proliferation of non-small-cell lung cancer.
Furthermore, by investigating the complex structure of the repebody with interleukin-6, Professor Kim has identified its mechanism, which demonstrated the potential for therapeutic development. The researchers are currently carrying out pre-clinical trials for acquiring permission to perform clinical trials on animals with non-small-cell lung cancer. The repebody can be developed into a new protein drug after demonstrating its safety and efficacy.
Professor Hak-Sung Kim and his team have confirmed that the repebody can be utilized as a new protein drug, and this will be a significant contribution to Korea’s protein drugs and biotechnology industry development.
The research was supported by the Future Pioneer Industry project and sponsored by the Ministry of Science, ICT and Future Planning.
Figure 1. Professor Kim’s article published as the cover article of July edition of Molecular Therapy
Figure 2. Clinical proof of the repebody’s inhibition of cancer growth using animal models
2014.07.14 View 13992 -
 Materials Developed for Sodium Rechargeable Battery by EEWS
The research group of Professor William Goddard III, You-Sung Jung, and Jang-Wook Choi from the Graduate School of Energy, Environment, Water, and Sustainability (EEWS) at KAIST has developed a new sodium-ion rechargeable battery which operates at a high voltage, can be charged, and stably discharges over 10,000 cycles. The research results were published in the online version of the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on December 30, 2013.
Since the material costs of sodium rechargeable batteries is 30 to 40 times lower than lithium batteries, it has received attention as an energy saving tool for smart grids and as the next generation of lithium rechargeable batteries. Until now, sodium-ion rechargeable batteries have had issues with stability when charging and discharging. The research group developed a vanadium-based electrode to solve these problems.
The group said follow-up research will be continued to develop advanced technology on sodium rechargeable batteries as it is still currently in the beginning stages.
The research team: From left to right is Professors William Goddard, You-Sung Jung, and Jang-Wook Choi
2014.01.13 View 11990
Materials Developed for Sodium Rechargeable Battery by EEWS
The research group of Professor William Goddard III, You-Sung Jung, and Jang-Wook Choi from the Graduate School of Energy, Environment, Water, and Sustainability (EEWS) at KAIST has developed a new sodium-ion rechargeable battery which operates at a high voltage, can be charged, and stably discharges over 10,000 cycles. The research results were published in the online version of the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on December 30, 2013.
Since the material costs of sodium rechargeable batteries is 30 to 40 times lower than lithium batteries, it has received attention as an energy saving tool for smart grids and as the next generation of lithium rechargeable batteries. Until now, sodium-ion rechargeable batteries have had issues with stability when charging and discharging. The research group developed a vanadium-based electrode to solve these problems.
The group said follow-up research will be continued to develop advanced technology on sodium rechargeable batteries as it is still currently in the beginning stages.
The research team: From left to right is Professors William Goddard, You-Sung Jung, and Jang-Wook Choi
2014.01.13 View 11990 -
 Quantum Mechanical Calculation Theory Developed
An Electron Density Functional Calculation Theory, based on the widely used quantum mechanical principles and yet accurate and with shortened calculation period, was developed by Korean research team.
*Electron Density Functional Calculation Theory: Theory that proves that it is possible to calculate energy and properties with only simple wave equations and electron densities.
The research was conducted by Professor Jeong Yoo Sung (Graduate School of EEWS) and Professor William Goddard with support from WCU Foster Project initiated by Ministry of Education, Science and Technology and Korea Research Foundation. The result was published in the Proceedings of the National Academy of Sciences Journal.
The research team corrected the error when performing quantum calculations that arises from the length of calculation time and incorrect assumptions and developed a theory and algorithm that is more accurate and faster. The use of wave equations in quantum mechanical calculations results in high accuracy but there is a rapid increase in calculation time and is therefore difficult to implement in large molecules with hundreds, or thousands of atoms. By implementing a low electron density variable with relatively less calculation work, the size of calculable molecule increases but the accuracy decreases.
The team focused on the interaction between electrons with different spins to improve upon the speed of calculation in the conventional accurate calculation. The team used the fact that the interaction between electrons with different spins increases as it comes closer together in accordance with the Pauli’s Exclusion Principle.
In addition the interaction between electrons are local and therefore can ignore the interactions between far away electrons and still get the total energy value. The team also took advantage of this fact and developed the algorithm that decreased calculation time hundredth fold.
Professor Jeong commented that, “So far most of the domestic achievements were made by focusing on integrative researches by calculation science and material design communities but these involved short time frames. In areas that required lengthy time frames like fundamentals and software development, there was no competitive advantage. However this research is significant in that a superior solution was developed domestically”.
2012.01.31 View 12547
Quantum Mechanical Calculation Theory Developed
An Electron Density Functional Calculation Theory, based on the widely used quantum mechanical principles and yet accurate and with shortened calculation period, was developed by Korean research team.
*Electron Density Functional Calculation Theory: Theory that proves that it is possible to calculate energy and properties with only simple wave equations and electron densities.
The research was conducted by Professor Jeong Yoo Sung (Graduate School of EEWS) and Professor William Goddard with support from WCU Foster Project initiated by Ministry of Education, Science and Technology and Korea Research Foundation. The result was published in the Proceedings of the National Academy of Sciences Journal.
The research team corrected the error when performing quantum calculations that arises from the length of calculation time and incorrect assumptions and developed a theory and algorithm that is more accurate and faster. The use of wave equations in quantum mechanical calculations results in high accuracy but there is a rapid increase in calculation time and is therefore difficult to implement in large molecules with hundreds, or thousands of atoms. By implementing a low electron density variable with relatively less calculation work, the size of calculable molecule increases but the accuracy decreases.
The team focused on the interaction between electrons with different spins to improve upon the speed of calculation in the conventional accurate calculation. The team used the fact that the interaction between electrons with different spins increases as it comes closer together in accordance with the Pauli’s Exclusion Principle.
In addition the interaction between electrons are local and therefore can ignore the interactions between far away electrons and still get the total energy value. The team also took advantage of this fact and developed the algorithm that decreased calculation time hundredth fold.
Professor Jeong commented that, “So far most of the domestic achievements were made by focusing on integrative researches by calculation science and material design communities but these involved short time frames. In areas that required lengthy time frames like fundamentals and software development, there was no competitive advantage. However this research is significant in that a superior solution was developed domestically”.
2012.01.31 View 12547 -
 Bioengineers develop a new strategy for accurate prediction of cellular metabolic fluxes
A team of pioneering South Korean scientists has developed a new strategy for accurately predicting cellular metabolic fluxes under various genotypic and environmental conditions. This groundbreaking research is published in the journal Proceedings of the National Academy of Sciences of the USA (PNAS) on August 2, 2010.
To understand cellular metabolism and predict its metabolic capability at systems-level, systems biological analysis by modeling and simulation of metabolic network plays an important role. The team from the Korea Advanced Institute of Science and Technology (KAIST), led by Distinguished Professor Sang Yup Lee, focused their research on the development of a new strategy for more accurate prediction of cellular metabolism.
“For strain improvement, biologists have made every effort to understand the global picture of biological systems and investigate the changes of all metabolic fluxes of the system under changing genotypic and environmental conditions,” said Lee. The accumulation of omics data, including genome, transcriptome, proteome, metabolome, and fluxome, provides an opportunity to understand the cellular physiology and metabolic characteristics at systems-level. With the availability of the fully annotated genome sequence, the genome-scale in silico (means “performed on computer or via computer simulation.”) metabolic models for a number of organisms have been successfully developed to improve our understanding on these biological systems. With these advances, the development of new simulation methods to analyze and integrate systematically large amounts of biological data and predict cellular metabolic capability for systems biological analysis is important.
Information used to reconstruct the genome-scale in silico cell is not yet complete, which can make the simulation results different from the physiological performances of the real cell. Thus, additional information and procedures, such as providing additional constraints (constraint: a term to exclude incorrect metabolic fluxes by restricting the solution space of in silico cell) to the model, are often incorporated to improve the accuracy of the in silico cell.
By employing information generated from the genome sequence and annotation, the KAIST team developed a new set of constraints, called Grouping Reaction (GR) constraints, to accurately predict metabolic fluxes. Based on the genomic information, functionally related reactions were organized into different groups. These groups were considered for the generation of GR constraints, as condition- and objective function- independent constraints. Since the method developed in this study does not require complex information but only the genome sequence and annotation, this strategy can be applied to any organism with a completely annotated genome sequence.
“As we become increasingly concerned with environmental problems and the limits of fossil resources, bio-based production of chemicals from renewable biomass has been receiving great attention. Systems biological analysis by modeling and simulation of biological systems, to understand cellular metabolism and identify the targets for the strain improvement, has provided a new paradigm for developing successful bioprocesses,” concluded Lee. This new strategy for predicting cellular metabolism is expected to contribute to more accurate determination of cellular metabolic characteristics, and consequently to the development of metabolic engineering strategies for the efficient production of important industrial products and identification of new drug targets in pathogens.”
2010.08.05 View 15532
Bioengineers develop a new strategy for accurate prediction of cellular metabolic fluxes
A team of pioneering South Korean scientists has developed a new strategy for accurately predicting cellular metabolic fluxes under various genotypic and environmental conditions. This groundbreaking research is published in the journal Proceedings of the National Academy of Sciences of the USA (PNAS) on August 2, 2010.
To understand cellular metabolism and predict its metabolic capability at systems-level, systems biological analysis by modeling and simulation of metabolic network plays an important role. The team from the Korea Advanced Institute of Science and Technology (KAIST), led by Distinguished Professor Sang Yup Lee, focused their research on the development of a new strategy for more accurate prediction of cellular metabolism.
“For strain improvement, biologists have made every effort to understand the global picture of biological systems and investigate the changes of all metabolic fluxes of the system under changing genotypic and environmental conditions,” said Lee. The accumulation of omics data, including genome, transcriptome, proteome, metabolome, and fluxome, provides an opportunity to understand the cellular physiology and metabolic characteristics at systems-level. With the availability of the fully annotated genome sequence, the genome-scale in silico (means “performed on computer or via computer simulation.”) metabolic models for a number of organisms have been successfully developed to improve our understanding on these biological systems. With these advances, the development of new simulation methods to analyze and integrate systematically large amounts of biological data and predict cellular metabolic capability for systems biological analysis is important.
Information used to reconstruct the genome-scale in silico cell is not yet complete, which can make the simulation results different from the physiological performances of the real cell. Thus, additional information and procedures, such as providing additional constraints (constraint: a term to exclude incorrect metabolic fluxes by restricting the solution space of in silico cell) to the model, are often incorporated to improve the accuracy of the in silico cell.
By employing information generated from the genome sequence and annotation, the KAIST team developed a new set of constraints, called Grouping Reaction (GR) constraints, to accurately predict metabolic fluxes. Based on the genomic information, functionally related reactions were organized into different groups. These groups were considered for the generation of GR constraints, as condition- and objective function- independent constraints. Since the method developed in this study does not require complex information but only the genome sequence and annotation, this strategy can be applied to any organism with a completely annotated genome sequence.
“As we become increasingly concerned with environmental problems and the limits of fossil resources, bio-based production of chemicals from renewable biomass has been receiving great attention. Systems biological analysis by modeling and simulation of biological systems, to understand cellular metabolism and identify the targets for the strain improvement, has provided a new paradigm for developing successful bioprocesses,” concluded Lee. This new strategy for predicting cellular metabolism is expected to contribute to more accurate determination of cellular metabolic characteristics, and consequently to the development of metabolic engineering strategies for the efficient production of important industrial products and identification of new drug targets in pathogens.”
2010.08.05 View 15532 -
 Native-like Spider Silk Produced in Metabolically Engineered Bacterium
Microscopic picture of 285 kilodalton recombinant spider silk fiber
Researchers have long envied spiders’ ability to manufacture silk that is light-weighted while as strong and tough as steel or Kevlar. Indeed, finer than human hair, five times stronger by weight than steel, and three times tougher than the top quality man-made fiber Kevlar, spider dragline silk is an ideal material for numerous applications. Suggested industrial applications have ranged from parachute cords and protective clothing to composite materials in aircrafts. Also, many biomedical applications are envisioned due to its biocompatibility and biodegradability.
Unfortunately, natural dragline silk cannot be conveniently obtained by farming spiders because they are highly territorial and aggressive. To develop a more sustainable process, can scientists mass-produce artificial silk while maintaining the amazing properties of native silk? That is something Sang Yup Lee at the Korea Advanced Institute of Science and Technology (KAIST) in Daejeon, the Republic of Korea, and his collaborators, Professor Young Hwan Park at Seoul National University and Professor David Kaplan at Tufts University, wanted to figure out. Their method is very similar to what spiders essentially do: first, expression of recombinant silk proteins; second, making the soluble silk proteins into water-insoluble fibers through spinning.
For the successful expression of high molecular weight spider silk protein, Professor Lee and his colleagues pieced together the silk gene from chemically synthesized oligonucleotides, and then inserted it into the expression host (in this case, an industrially safe bacterium Escherichia coli which is normally found in our gut). Initially, the bacterium refused to the challenging task of producing high molecular weight spider silk protein due to the unique characteristics of the protein, such as extremely large size, repetitive nature of the protein structure, and biased abundance of a particular amino acid glycine. “To make E. coli synthesize this ultra high molecular weight (as big as 285 kilodalton) spider silk protein having highly repetitive amino acid sequence, we helped E. coli overcome the difficulties by systems metabolic engineering,” says Sang Yup Lee, Distinguished Professor of KAIST, who led this project. His team boosted the pool of glycyl-tRNA, the major building block of spider silk protein synthesis. “We could obtain appreciable expression of the 285 kilodalton spider silk protein, which is the largest recombinant silk protein ever produced in E. coli. That was really incredible.” says Dr. Xia.
But this was only step one. The KAIST team performed high-cell-density cultures for mass production of the recombinant spider silk protein. Then, the team developed a simple, easy to scale-up purification process for the recombinant spider silk protein. The purified spider silk protein could be spun into beautiful silk fiber. To study the mechanical properties of the artificial spider silk, the researchers determined tenacity, elongation, and Young’s modulus, the three critical mechanical parameters that represent a fiber’s strength, extensibility, and stiffness. Importantly, the artificial fiber displayed the tenacity, elongation, and Young’s modulus of 508 MPa, 15%, and 21 GPa, respectively, which are comparable to those of the native spider silk.
“We have offered an overall platform for mass production of native-like spider dragline silk. This platform would enable us to have broader industrial and biomedical applications for spider silk. Moreover, many other silk-like biomaterials such as elastin, collagen, byssus, resilin, and other repetitive proteins have similar features to spider silk protein. Thus, our platform should also be useful for their efficient bio-based production and applications,” concludes Professor Lee.
This work is published on July 26 in the Proceedings of the National Academy of Sciences (PNAS) online.
2010.07.28 View 19912
Native-like Spider Silk Produced in Metabolically Engineered Bacterium
Microscopic picture of 285 kilodalton recombinant spider silk fiber
Researchers have long envied spiders’ ability to manufacture silk that is light-weighted while as strong and tough as steel or Kevlar. Indeed, finer than human hair, five times stronger by weight than steel, and three times tougher than the top quality man-made fiber Kevlar, spider dragline silk is an ideal material for numerous applications. Suggested industrial applications have ranged from parachute cords and protective clothing to composite materials in aircrafts. Also, many biomedical applications are envisioned due to its biocompatibility and biodegradability.
Unfortunately, natural dragline silk cannot be conveniently obtained by farming spiders because they are highly territorial and aggressive. To develop a more sustainable process, can scientists mass-produce artificial silk while maintaining the amazing properties of native silk? That is something Sang Yup Lee at the Korea Advanced Institute of Science and Technology (KAIST) in Daejeon, the Republic of Korea, and his collaborators, Professor Young Hwan Park at Seoul National University and Professor David Kaplan at Tufts University, wanted to figure out. Their method is very similar to what spiders essentially do: first, expression of recombinant silk proteins; second, making the soluble silk proteins into water-insoluble fibers through spinning.
For the successful expression of high molecular weight spider silk protein, Professor Lee and his colleagues pieced together the silk gene from chemically synthesized oligonucleotides, and then inserted it into the expression host (in this case, an industrially safe bacterium Escherichia coli which is normally found in our gut). Initially, the bacterium refused to the challenging task of producing high molecular weight spider silk protein due to the unique characteristics of the protein, such as extremely large size, repetitive nature of the protein structure, and biased abundance of a particular amino acid glycine. “To make E. coli synthesize this ultra high molecular weight (as big as 285 kilodalton) spider silk protein having highly repetitive amino acid sequence, we helped E. coli overcome the difficulties by systems metabolic engineering,” says Sang Yup Lee, Distinguished Professor of KAIST, who led this project. His team boosted the pool of glycyl-tRNA, the major building block of spider silk protein synthesis. “We could obtain appreciable expression of the 285 kilodalton spider silk protein, which is the largest recombinant silk protein ever produced in E. coli. That was really incredible.” says Dr. Xia.
But this was only step one. The KAIST team performed high-cell-density cultures for mass production of the recombinant spider silk protein. Then, the team developed a simple, easy to scale-up purification process for the recombinant spider silk protein. The purified spider silk protein could be spun into beautiful silk fiber. To study the mechanical properties of the artificial spider silk, the researchers determined tenacity, elongation, and Young’s modulus, the three critical mechanical parameters that represent a fiber’s strength, extensibility, and stiffness. Importantly, the artificial fiber displayed the tenacity, elongation, and Young’s modulus of 508 MPa, 15%, and 21 GPa, respectively, which are comparable to those of the native spider silk.
“We have offered an overall platform for mass production of native-like spider dragline silk. This platform would enable us to have broader industrial and biomedical applications for spider silk. Moreover, many other silk-like biomaterials such as elastin, collagen, byssus, resilin, and other repetitive proteins have similar features to spider silk protein. Thus, our platform should also be useful for their efficient bio-based production and applications,” concludes Professor Lee.
This work is published on July 26 in the Proceedings of the National Academy of Sciences (PNAS) online.
2010.07.28 View 19912