Nature
-
 KAIST Predicts Diseases by Early Detection of Aging Signals in Liver Tissue
- KAIST-KRIBB Develops ‘FiNi-seq’ Technology to Capture Characteristics of Fibrotic Microenvironments Accumulated in Liver Tissue and Dynamic Changes of Early Aging Cells
- Elucidation of the Spatial Ecosystem of Aged Liver Tissue, where Reprogramming of Senescent Cells and Immune Exhaustion Progresses, at the Single-Cell Genome and Epigenome Levels
< (From left) Professor Jong-Eun Park of KAIST Graduate School of Medical Science and Engineering (GSMSE), Dr. Chuna Kim of KRIBB, Dr. Kwon Yong Tak of KAIST GSMSE, Ph.D. Candidate Juyeon Kim of KRIBB, Ph.D. Candidate Myungsun Park of KAIST GSMSE >
Aging and chronic diseases involve the gradual accumulation of subtle tissue changes over a long period. Therefore, there are still limitations in quantitatively understanding these changes within organs and linking them to early signs of disease onset. In response, Korean researchers have successfully developed a platform technology that accurately captures localized changes that first occur within tissue, significantly aiding in faster disease discovery and prediction, and in setting personalized treatment targets.
KAIST (President Kwang Hyung Lee) announced on June 12th that a joint research team led by Professor Jong-Eun Park of the Graduate School of Medical Science and Engineering at KAIST and Dr. Chuna Kim of the Aging Convergence Research Center at the Korea Research Institute of Bioscience and Biotechnology (KRIBB, President Seok-Yoon Kwon) has developed ‘FiNi-seq (Fibrotic Niche enrichment sequencing)’ technology. This technology captures fibrotic microenvironments locally occurring in aged liver tissue and enables precise analysis at the single-cell transcriptome level*.
*Single-cell transcriptome analysis: A method to measure how actively each cell uses which genes, allowing identification and function of individual diseased cells.
The researchers developed a method to selectively enrich early aging microenvironments where regeneration is delayed and fibrosis accumulates, by physically selecting regions with high tissue degradation resistance in aged liver tissue.
In this process, high-resolution identification of fibrosis-related endothelial cells, fibroblasts interacting with the immune system, and immune-exhausted cells such as PD-1 highly expressing CD8 T cells, which were difficult to capture with existing single-cell analysis technologies, was possible.
In particular, the research team confirmed through ‘FiNi-seq’ technology that specific cells observed in fibrotic areas within aged liver tissue secondarily age the surrounding environment through secreted factors, and that this leads to the expansion of the aged environment.
Furthermore, they also elucidated the mechanism by which endothelial cells lose their tissue-specific identity and induce innate immune responses, promoting immune cell infiltration. Through spatial transcriptome analysis, the spatial distribution of fibroblasts interacting with immune cells was quantified, revealing their involvement in tissue regeneration, induction of inflammatory responses, and progression to chronic fibrosis.
The research team performed integrated analysis of multi-omics\* data to obtain transcriptome and epigenome information, precisely interpreting the microenvironment of aged liver tissue and its spatial heterogeneity, and confirming how these changes are connected to the intrahepatic vascular structure.
*Multi-omics: An integrated analysis method for various biological information within an organism, such as genes, proteins, metabolites, and cell information.
The newly developed ‘FiNi-seq’ technology is expected to be a useful platform for high-resolution capture of pathophysiological signals in most chronic liver diseases, including the aging process that causes fibrosis.
< Figure 1. Isolation of fibrotic regions from aged liver tissue, followed by single-cell transcriptome analysis and validation in a fibrosis model. >
The first author, Dr. Kwon Yong Tak of KAIST Graduate School of Medical Science and Engineering (GSMSE), a hepatologist at Seoul St. Mary's Hospital, designed this study to lay the groundwork for early diagnosis and treatment of fibrosis progression, the most important clinical prognostic indicator in chronic liver disease, while pursuing his Ph.D. at KAIST KAIST GSMSE with support from the physician-scientist training program. Co-first author Myungsun Park, a Ph.D. candidate at KAIST KAIST GSMSE, was responsible for the technical implementation of FiNi-seq technology, and Juyeon Kim, a Ph.D. candidate at KRIBB's Aging Convergence Research Center, was responsible for imaging analysis of aged tissue, playing a key role in the research.
Dr. Chuna Kim of KRIBB stated, “Through this study, we were able to precisely elucidate the cellular composition and spatial characteristics of the fibrotic microenvironment observed in aged liver tissue at the single-cell level.”
< Figure 2. Spatially defined stepwise progression patterns of aging-related regions within the liver and identification of regulatory factors inducing them. >
Professor Jong-Eun Park of the Graduate School of Medical Science and Engineering said, “As an analytical technology that can capture subtle changes occurring in the early stages of aging and chronic diseases, it is expected to play a significant role in finding effective treatment targets in the future. Also, we plan to expand this research to chronic diseases in other organs such as the lungs and kidneys, as well as various liver disease models.”
This research was published in the international journal ‘Nature Aging’ on May 5, 2025, with Dr. Kwon Yong Tak of KAIST KAIST GSMSE, Ph.D. Candidate Juyeon Kim of KRIBB, and Ph.D. Candidate Myungsun Park of KAIST as co-first authors.
*Paper Title: Quasi-spatial single-cell transcriptome based on physical tissue properties defines early aging associated niche in liver
*DOI: https://doi.org/10.1038/s43587-025-00857-7
This research was supported by several domestic institutions, including the National Research Foundation of Korea, the Korea Health Industry Development Institute (KHIDI), the Korea Research Institute of Bioscience and Biotechnology (KRIBB), KIST, POSCO Science Fellowship, and the Convergence Medical Scientist Training Program.
2025.06.12 View 177
KAIST Predicts Diseases by Early Detection of Aging Signals in Liver Tissue
- KAIST-KRIBB Develops ‘FiNi-seq’ Technology to Capture Characteristics of Fibrotic Microenvironments Accumulated in Liver Tissue and Dynamic Changes of Early Aging Cells
- Elucidation of the Spatial Ecosystem of Aged Liver Tissue, where Reprogramming of Senescent Cells and Immune Exhaustion Progresses, at the Single-Cell Genome and Epigenome Levels
< (From left) Professor Jong-Eun Park of KAIST Graduate School of Medical Science and Engineering (GSMSE), Dr. Chuna Kim of KRIBB, Dr. Kwon Yong Tak of KAIST GSMSE, Ph.D. Candidate Juyeon Kim of KRIBB, Ph.D. Candidate Myungsun Park of KAIST GSMSE >
Aging and chronic diseases involve the gradual accumulation of subtle tissue changes over a long period. Therefore, there are still limitations in quantitatively understanding these changes within organs and linking them to early signs of disease onset. In response, Korean researchers have successfully developed a platform technology that accurately captures localized changes that first occur within tissue, significantly aiding in faster disease discovery and prediction, and in setting personalized treatment targets.
KAIST (President Kwang Hyung Lee) announced on June 12th that a joint research team led by Professor Jong-Eun Park of the Graduate School of Medical Science and Engineering at KAIST and Dr. Chuna Kim of the Aging Convergence Research Center at the Korea Research Institute of Bioscience and Biotechnology (KRIBB, President Seok-Yoon Kwon) has developed ‘FiNi-seq (Fibrotic Niche enrichment sequencing)’ technology. This technology captures fibrotic microenvironments locally occurring in aged liver tissue and enables precise analysis at the single-cell transcriptome level*.
*Single-cell transcriptome analysis: A method to measure how actively each cell uses which genes, allowing identification and function of individual diseased cells.
The researchers developed a method to selectively enrich early aging microenvironments where regeneration is delayed and fibrosis accumulates, by physically selecting regions with high tissue degradation resistance in aged liver tissue.
In this process, high-resolution identification of fibrosis-related endothelial cells, fibroblasts interacting with the immune system, and immune-exhausted cells such as PD-1 highly expressing CD8 T cells, which were difficult to capture with existing single-cell analysis technologies, was possible.
In particular, the research team confirmed through ‘FiNi-seq’ technology that specific cells observed in fibrotic areas within aged liver tissue secondarily age the surrounding environment through secreted factors, and that this leads to the expansion of the aged environment.
Furthermore, they also elucidated the mechanism by which endothelial cells lose their tissue-specific identity and induce innate immune responses, promoting immune cell infiltration. Through spatial transcriptome analysis, the spatial distribution of fibroblasts interacting with immune cells was quantified, revealing their involvement in tissue regeneration, induction of inflammatory responses, and progression to chronic fibrosis.
The research team performed integrated analysis of multi-omics\* data to obtain transcriptome and epigenome information, precisely interpreting the microenvironment of aged liver tissue and its spatial heterogeneity, and confirming how these changes are connected to the intrahepatic vascular structure.
*Multi-omics: An integrated analysis method for various biological information within an organism, such as genes, proteins, metabolites, and cell information.
The newly developed ‘FiNi-seq’ technology is expected to be a useful platform for high-resolution capture of pathophysiological signals in most chronic liver diseases, including the aging process that causes fibrosis.
< Figure 1. Isolation of fibrotic regions from aged liver tissue, followed by single-cell transcriptome analysis and validation in a fibrosis model. >
The first author, Dr. Kwon Yong Tak of KAIST Graduate School of Medical Science and Engineering (GSMSE), a hepatologist at Seoul St. Mary's Hospital, designed this study to lay the groundwork for early diagnosis and treatment of fibrosis progression, the most important clinical prognostic indicator in chronic liver disease, while pursuing his Ph.D. at KAIST KAIST GSMSE with support from the physician-scientist training program. Co-first author Myungsun Park, a Ph.D. candidate at KAIST KAIST GSMSE, was responsible for the technical implementation of FiNi-seq technology, and Juyeon Kim, a Ph.D. candidate at KRIBB's Aging Convergence Research Center, was responsible for imaging analysis of aged tissue, playing a key role in the research.
Dr. Chuna Kim of KRIBB stated, “Through this study, we were able to precisely elucidate the cellular composition and spatial characteristics of the fibrotic microenvironment observed in aged liver tissue at the single-cell level.”
< Figure 2. Spatially defined stepwise progression patterns of aging-related regions within the liver and identification of regulatory factors inducing them. >
Professor Jong-Eun Park of the Graduate School of Medical Science and Engineering said, “As an analytical technology that can capture subtle changes occurring in the early stages of aging and chronic diseases, it is expected to play a significant role in finding effective treatment targets in the future. Also, we plan to expand this research to chronic diseases in other organs such as the lungs and kidneys, as well as various liver disease models.”
This research was published in the international journal ‘Nature Aging’ on May 5, 2025, with Dr. Kwon Yong Tak of KAIST KAIST GSMSE, Ph.D. Candidate Juyeon Kim of KRIBB, and Ph.D. Candidate Myungsun Park of KAIST as co-first authors.
*Paper Title: Quasi-spatial single-cell transcriptome based on physical tissue properties defines early aging associated niche in liver
*DOI: https://doi.org/10.1038/s43587-025-00857-7
This research was supported by several domestic institutions, including the National Research Foundation of Korea, the Korea Health Industry Development Institute (KHIDI), the Korea Research Institute of Bioscience and Biotechnology (KRIBB), KIST, POSCO Science Fellowship, and the Convergence Medical Scientist Training Program.
2025.06.12 View 177 -
 KAIST develops technology for selective RNA modification in living cells and animals
· A team led by Professor Won Do Heo from the Department of Biological Sciences, KAIST, has developed a pioneering technology that selectively acetylates specific RNA molecules in living cells and tissues.
· The platform uses RNA-targeting CRISPR tools in combination with RNA-modifying enzymes to chemically modify only the intended RNA.
· The method opens new possibilities for gene therapy by enabling precise control of disease-related RNA without affecting the rest of the transcriptome.
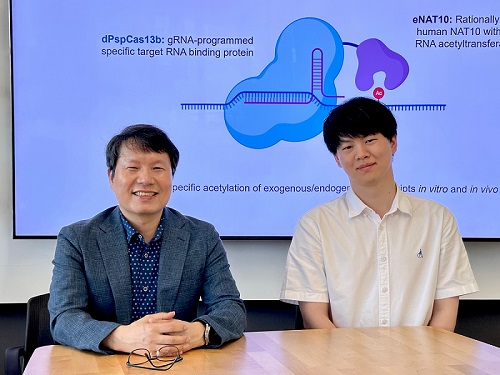
< Photo 1. (From left) Professor Won Do Heo and Jihwan Yu, a Ph.D. Candidate of the Department of Biological Sciences >
CRISPR-Cas13, a powerful RNA-targeting technology is gaining increasing attention as a next-generation gene therapy platform due to its precision and reduced side effects. Utilizing this system, researchers at KAIST have now developed the world’s first technology capable of selectively acetylating (chemically modifying) specific RNA molecules among countless transcripts within living cells. This breakthrough enables precise, programmable control of RNA function and is expected to open new avenues in RNA-based therapeutic development.
KAIST (President Kwang Hyung Lee) announced that a research team led by Professor Won Do Heo in the Department of Biological Sciences has recently developed a groundbreaking technology capable of selectively acetylating specific RNA molecules within the human body using the CRISPR-Cas13 system—an RNA-targeting platform gaining increasing attention in the fields of gene regulation and RNA-based therapeutics.
RNA molecules can undergo chemical modifications—the addition of specific chemical groups—which alter their function and behavior without changing the underlying nucleotide sequence. However, some of these modifications, a critical layer of post-transcriptional gene regulation, remain poorly understood. Among them, N4-acetylcytidine (ac4C) has been particularly enigmatic, with ongoing debate about its existence and function in human messenger RNA (mRNA), the RNA that encodes proteins.
To address this gap, the KAIST research team developed a targeted RNA acetylation system, named dCas13-eNAT10. This platform combines a catalytically inactive Cas13 enzyme (dCas13) that guides the system to specific RNA targets, with a hyperactive variant of the NAT10 enzyme (eNAT10), which performs RNA acetylation. This approach enables precise acetylation of only the desired RNA molecules among the vast pool of transcripts within the cell.
< Figure 1. Development of hyperactive variant eNAT10 through NAT10 protein engineering. By engineering the NAT10 protein, which performs RNA acetylation in human cells, based on its domain and structure, eNAT10 was developed, showing approximately a 3-fold increase in RNA acetylation activity compared to the wild-type enzyme. >
Using this system, the researchers demonstrated that guide RNAs could direct the dCas13-eNAT10 complex to acetylate specific RNA targets, and acetylation significantly increased protein expression from the modified mRNA. Moreover, the study revealed, for the first time, that RNA acetylation plays a role in intracellular RNA localization, facilitating the export of RNA from the nucleus to the cytoplasm—a critical step in gene expression regulation.
To validate its therapeutic potential, the team successfully delivered the targeted RNA acetylation system into the livers of live mice using adeno-associated virus (AAV), a commonly used gene therapy vector. This marks the first demonstration of in vivo RNA modification, extending the applicability of RNA chemical modification tools from cell culture models to living organisms.
< Figure 2. Acetylation of various RNA in cells using dCas13-eNAT10 fusion protein. Utilizing the CRISPR-Cas13 system, which can precisely target specific RNA through guide RNA, a dCas13-eNAT10 fusion protein was created, demonstrating its ability to specifically acetylate various endogenous RNA at different locations within cells. >
Professor Won Do Heo, who previously developed COVID-19 treatment technology using RNA gene scissors and technology to activate RNA gene scissors with light, stated, "Existing RNA chemical modification research faced difficulties in controlling specificity, temporality, and spatiality. However, this new technology allows selective acetylation of desired RNA, opening the door for accurate and detailed research into the functions of RNA acetylation." He added, "The RNA chemical modification technology developed in this study can be widely used as an RNA-based therapeutic agent and a tool for regulating RNA functions in living organisms in the future."
< Figure 3. In vivo delivery of targeted RNA acetylation system. The targeted RNA acetylation system was encoded in an AAV vector, commonly used in gene therapy, and delivered intravenously to adult mice, showing that target RNA in liver tissue was specifically acetylated according to the guide RNA. >
This research, with Ph.D. candidate Jihwan Yu from the Department of Biological Sciences at KAIST as the first author, was published in the journal Nature Chemical Biology on June 2, 2025. (Title: Programmable RNA acetylation with CRISPR-Cas13, Impact factor: 12.9, DOI: https://doi.org/10.1038/s41589-025-01922-3)
This research was supported by the Samsung Future Technology Foundation and the Bio & Medical Technology Development Program of the National Research Foundation of Korea.
2025.06.10 View 270
KAIST develops technology for selective RNA modification in living cells and animals
· A team led by Professor Won Do Heo from the Department of Biological Sciences, KAIST, has developed a pioneering technology that selectively acetylates specific RNA molecules in living cells and tissues.
· The platform uses RNA-targeting CRISPR tools in combination with RNA-modifying enzymes to chemically modify only the intended RNA.
· The method opens new possibilities for gene therapy by enabling precise control of disease-related RNA without affecting the rest of the transcriptome.
< Photo 1. (From left) Professor Won Do Heo and Jihwan Yu, a Ph.D. Candidate of the Department of Biological Sciences >
CRISPR-Cas13, a powerful RNA-targeting technology is gaining increasing attention as a next-generation gene therapy platform due to its precision and reduced side effects. Utilizing this system, researchers at KAIST have now developed the world’s first technology capable of selectively acetylating (chemically modifying) specific RNA molecules among countless transcripts within living cells. This breakthrough enables precise, programmable control of RNA function and is expected to open new avenues in RNA-based therapeutic development.
KAIST (President Kwang Hyung Lee) announced that a research team led by Professor Won Do Heo in the Department of Biological Sciences has recently developed a groundbreaking technology capable of selectively acetylating specific RNA molecules within the human body using the CRISPR-Cas13 system—an RNA-targeting platform gaining increasing attention in the fields of gene regulation and RNA-based therapeutics.
RNA molecules can undergo chemical modifications—the addition of specific chemical groups—which alter their function and behavior without changing the underlying nucleotide sequence. However, some of these modifications, a critical layer of post-transcriptional gene regulation, remain poorly understood. Among them, N4-acetylcytidine (ac4C) has been particularly enigmatic, with ongoing debate about its existence and function in human messenger RNA (mRNA), the RNA that encodes proteins.
To address this gap, the KAIST research team developed a targeted RNA acetylation system, named dCas13-eNAT10. This platform combines a catalytically inactive Cas13 enzyme (dCas13) that guides the system to specific RNA targets, with a hyperactive variant of the NAT10 enzyme (eNAT10), which performs RNA acetylation. This approach enables precise acetylation of only the desired RNA molecules among the vast pool of transcripts within the cell.
< Figure 1. Development of hyperactive variant eNAT10 through NAT10 protein engineering. By engineering the NAT10 protein, which performs RNA acetylation in human cells, based on its domain and structure, eNAT10 was developed, showing approximately a 3-fold increase in RNA acetylation activity compared to the wild-type enzyme. >
Using this system, the researchers demonstrated that guide RNAs could direct the dCas13-eNAT10 complex to acetylate specific RNA targets, and acetylation significantly increased protein expression from the modified mRNA. Moreover, the study revealed, for the first time, that RNA acetylation plays a role in intracellular RNA localization, facilitating the export of RNA from the nucleus to the cytoplasm—a critical step in gene expression regulation.
To validate its therapeutic potential, the team successfully delivered the targeted RNA acetylation system into the livers of live mice using adeno-associated virus (AAV), a commonly used gene therapy vector. This marks the first demonstration of in vivo RNA modification, extending the applicability of RNA chemical modification tools from cell culture models to living organisms.
< Figure 2. Acetylation of various RNA in cells using dCas13-eNAT10 fusion protein. Utilizing the CRISPR-Cas13 system, which can precisely target specific RNA through guide RNA, a dCas13-eNAT10 fusion protein was created, demonstrating its ability to specifically acetylate various endogenous RNA at different locations within cells. >
Professor Won Do Heo, who previously developed COVID-19 treatment technology using RNA gene scissors and technology to activate RNA gene scissors with light, stated, "Existing RNA chemical modification research faced difficulties in controlling specificity, temporality, and spatiality. However, this new technology allows selective acetylation of desired RNA, opening the door for accurate and detailed research into the functions of RNA acetylation." He added, "The RNA chemical modification technology developed in this study can be widely used as an RNA-based therapeutic agent and a tool for regulating RNA functions in living organisms in the future."
< Figure 3. In vivo delivery of targeted RNA acetylation system. The targeted RNA acetylation system was encoded in an AAV vector, commonly used in gene therapy, and delivered intravenously to adult mice, showing that target RNA in liver tissue was specifically acetylated according to the guide RNA. >
This research, with Ph.D. candidate Jihwan Yu from the Department of Biological Sciences at KAIST as the first author, was published in the journal Nature Chemical Biology on June 2, 2025. (Title: Programmable RNA acetylation with CRISPR-Cas13, Impact factor: 12.9, DOI: https://doi.org/10.1038/s41589-025-01922-3)
This research was supported by the Samsung Future Technology Foundation and the Bio & Medical Technology Development Program of the National Research Foundation of Korea.
2025.06.10 View 270 -
 KAIST Develops Virtual Staining Technology for 3D Histopathology
Moving beyond traditional methods of observing thinly sliced and stained cancer tissues, a collaborative international research team led by KAIST has successfully developed a groundbreaking technology. This innovation uses advanced optical techniques combined with an artificial intelligence-based deep learning algorithm to create realistic, virtually stained 3D images of cancer tissue without the need for serial sectioning nor staining. This breakthrough is anticipated to pave the way for next-generation non-invasive pathological diagnosis.
< Photo 1. (From left) Juyeon Park (Ph.D. Candidate, Department of Physics), Professor YongKeun Park (Department of Physics) (Top left) Professor Su-Jin Shin (Gangnam Severance Hospital), Professor Tae Hyun Hwang (Vanderbilt University School of Medicine) >
KAIST (President Kwang Hyung Lee) announced on the 26th that a research team led by Professor YongKeun Park of the Department of Physics, in collaboration with Professor Su-Jin Shin's team at Yonsei University Gangnam Severance Hospital, Professor Tae Hyun Hwang's team at Mayo Clinic, and Tomocube's AI research team, has developed an innovative technology capable of vividly displaying the 3D structure of cancer tissues without separate staining.
For over 200 years, conventional pathology has relied on observing cancer tissues under a microscope, a method that only shows specific cross-sections of the 3D cancer tissue. This has limited the ability to understand the three-dimensional connections and spatial arrangements between cells.
To overcome this, the research team utilized holotomography (HT), an advanced optical technology, to measure the 3D refractive index information of tissues. They then integrated an AI-based deep learning algorithm to successfully generate virtual H&E* images.* H&E (Hematoxylin & Eosin): The most widely used staining method for observing pathological tissues. Hematoxylin stains cell nuclei blue, and eosin stains cytoplasm pink.
The research team quantitatively demonstrated that the images generated by this technology are highly similar to actual stained tissue images. Furthermore, the technology exhibited consistent performance across various organs and tissues, proving its versatility and reliability as a next-generation pathological analysis tool.
< Figure 1. Comparison of conventional 3D tissue pathology procedure and the 3D virtual H&E staining technology proposed in this study. The traditional method requires preparing and staining dozens of tissue slides, while the proposed technology can reduce the number of slides by up to 10 times and quickly generate H&E images without the staining process. >
Moreover, by validating the feasibility of this technology through joint research with hospitals and research institutions in Korea and the United States, utilizing Tomocube's holotomography equipment, the team demonstrated its potential for full-scale adoption in real-world pathological research settings.
Professor YongKeun Park stated, "This research marks a major advancement by transitioning pathological analysis from conventional 2D methods to comprehensive 3D imaging. It will greatly enhance biomedical research and clinical diagnostics, particularly in understanding cancer tumor boundaries and the intricate spatial arrangements of cells within tumor microenvironments."
< Figure 2. Results of AI-based 3D virtual H&E staining and quantitative analysis of pathological tissue. The virtually stained images enabled 3D reconstruction of key pathological features such as cell nuclei and glandular lumens. Based on this, various quantitative indicators, including cell nuclear distribution, volume, and surface area, could be extracted. >
This research, with Juyeon Park, a student of the Integrated Master’s and Ph.D. Program at KAIST, as the first author, was published online in the prestigious journal Nature Communications on May 22.
(Paper title: Revealing 3D microanatomical structures of unlabeled thick cancer tissues using holotomography and virtual H&E staining.
[https://doi.org/10.1038/s41467-025-59820-0]
This study was supported by the Leader Researcher Program of the National Research Foundation of Korea, the Global Industry Technology Cooperation Center Project of the Korea Institute for Advancement of Technology, and the Korea Health Industry Development Institute.
2025.05.26 View 1232
KAIST Develops Virtual Staining Technology for 3D Histopathology
Moving beyond traditional methods of observing thinly sliced and stained cancer tissues, a collaborative international research team led by KAIST has successfully developed a groundbreaking technology. This innovation uses advanced optical techniques combined with an artificial intelligence-based deep learning algorithm to create realistic, virtually stained 3D images of cancer tissue without the need for serial sectioning nor staining. This breakthrough is anticipated to pave the way for next-generation non-invasive pathological diagnosis.
< Photo 1. (From left) Juyeon Park (Ph.D. Candidate, Department of Physics), Professor YongKeun Park (Department of Physics) (Top left) Professor Su-Jin Shin (Gangnam Severance Hospital), Professor Tae Hyun Hwang (Vanderbilt University School of Medicine) >
KAIST (President Kwang Hyung Lee) announced on the 26th that a research team led by Professor YongKeun Park of the Department of Physics, in collaboration with Professor Su-Jin Shin's team at Yonsei University Gangnam Severance Hospital, Professor Tae Hyun Hwang's team at Mayo Clinic, and Tomocube's AI research team, has developed an innovative technology capable of vividly displaying the 3D structure of cancer tissues without separate staining.
For over 200 years, conventional pathology has relied on observing cancer tissues under a microscope, a method that only shows specific cross-sections of the 3D cancer tissue. This has limited the ability to understand the three-dimensional connections and spatial arrangements between cells.
To overcome this, the research team utilized holotomography (HT), an advanced optical technology, to measure the 3D refractive index information of tissues. They then integrated an AI-based deep learning algorithm to successfully generate virtual H&E* images.* H&E (Hematoxylin & Eosin): The most widely used staining method for observing pathological tissues. Hematoxylin stains cell nuclei blue, and eosin stains cytoplasm pink.
The research team quantitatively demonstrated that the images generated by this technology are highly similar to actual stained tissue images. Furthermore, the technology exhibited consistent performance across various organs and tissues, proving its versatility and reliability as a next-generation pathological analysis tool.
< Figure 1. Comparison of conventional 3D tissue pathology procedure and the 3D virtual H&E staining technology proposed in this study. The traditional method requires preparing and staining dozens of tissue slides, while the proposed technology can reduce the number of slides by up to 10 times and quickly generate H&E images without the staining process. >
Moreover, by validating the feasibility of this technology through joint research with hospitals and research institutions in Korea and the United States, utilizing Tomocube's holotomography equipment, the team demonstrated its potential for full-scale adoption in real-world pathological research settings.
Professor YongKeun Park stated, "This research marks a major advancement by transitioning pathological analysis from conventional 2D methods to comprehensive 3D imaging. It will greatly enhance biomedical research and clinical diagnostics, particularly in understanding cancer tumor boundaries and the intricate spatial arrangements of cells within tumor microenvironments."
< Figure 2. Results of AI-based 3D virtual H&E staining and quantitative analysis of pathological tissue. The virtually stained images enabled 3D reconstruction of key pathological features such as cell nuclei and glandular lumens. Based on this, various quantitative indicators, including cell nuclear distribution, volume, and surface area, could be extracted. >
This research, with Juyeon Park, a student of the Integrated Master’s and Ph.D. Program at KAIST, as the first author, was published online in the prestigious journal Nature Communications on May 22.
(Paper title: Revealing 3D microanatomical structures of unlabeled thick cancer tissues using holotomography and virtual H&E staining.
[https://doi.org/10.1038/s41467-025-59820-0]
This study was supported by the Leader Researcher Program of the National Research Foundation of Korea, the Global Industry Technology Cooperation Center Project of the Korea Institute for Advancement of Technology, and the Korea Health Industry Development Institute.
2025.05.26 View 1232 -
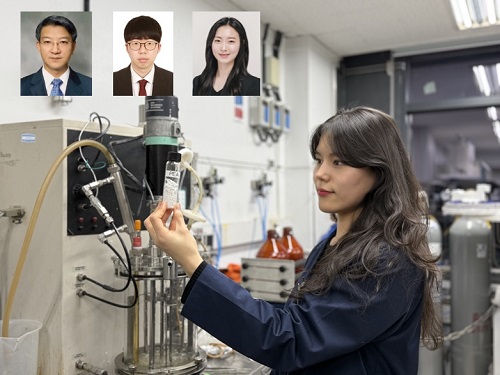 KAIST Develops Eco-Friendly, Nylon-Like Plastic Using Microorganisms
Poly(ester amide) amide is a next-generation material that combines the advantages of PET (polyester) and nylon (polyamide), two widely used plastics. However, it could only be produced from fossil fuels, which posed environmental concerns. Using microorganisms, KAIST researchers have successfully developed a new bio-based plastic to replace conventional plastic.
KAIST (represented by President Kwang Hyung Lee) announced on the 20th of March that a research team led by Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering has developed microbial strains through systems metabolic engineering to produce various eco-friendly, bio-based poly(ester amide)s. The team collaborated with researchers from the Korea Research Institute of Chemical Technology (KRICT, President Young-Kook Lee) to analyze and confirm the properties of the resulting plastic.
Professor Sang Yup Lee’s research team designed new metabolic pathways that do not naturally exist in microorganisms, and developed a platform microbial strain capable of producing nine different types of poly(ester amide)s, including poly(3-hydroxybutyrate-ran-3-aminopropionate) and poly(3-hydroxybutyrate-ran-4-aminobutyrate).
Using glucose derived from abundant biomass sources such as waste wood and weeds, the team successfully produced poly(ester amide)s in an eco-friendly manner. The researchers also confirmed the potential for industrial-scale production by demonstrating high production efficiency (54.57 g/L) using fed-batch fermentation of the engineered strain.
In collaboration with researchers Haemin Jeong and Jihoon Shin from KRICT, the KAIST team analyzed the properties of the bio-based plastic and found that it exhibited characteristics similar to high-density polyethylene (HDPE). This means the new plastic is not only eco-friendly but also strong and durable enough to replace conventional plastics.
The engineered strains and strategies developed in this study are expected to be useful not only for producing various poly(ester amide)s but also for constructing metabolic pathways for the biosynthesis of other types of polymers.
Professor Sang Yup Lee stated, “This study is the first to demonstrate the possibility of producing poly(ester amide)s (plastics) through a renewable bio-based chemical process rather than relying on the petroleum-based chemical industry. We plan to further enhance the production yield and efficiency through continued research.”
The study was published online on March 17 in the international journal Nature Chemical Biology.
·Title: Biosynthesis of poly(ester amide)s in engineered Escherichia coli
·DOI: 10.1038/s41589-025-01842-2
·Authors: A total of seven authors including Tong Un Chae (KAIST, first author), So Young Choi (KAIST, second author), Da-Hee Ahn (KAIST, third author), Woo Dae Jang (KAIST, fourth author), Haemin Jeong (KRICT, fifth author), Jihoon Shin (KRICT, sixth author), and Sang Yup Lee (KAIST, corresponding author).
This research was supported by the Ministry of Science and ICT (MSIT) under the Eco-Friendly Chemical Technology Development Project as part of the "Next-Generation Biorefinery Technology Development to Lead the Bio-Chemical Industry" initiative (project led by Distinguished Professor Sang Yup Lee at KAIST).
2025.03.24 View 3694
KAIST Develops Eco-Friendly, Nylon-Like Plastic Using Microorganisms
Poly(ester amide) amide is a next-generation material that combines the advantages of PET (polyester) and nylon (polyamide), two widely used plastics. However, it could only be produced from fossil fuels, which posed environmental concerns. Using microorganisms, KAIST researchers have successfully developed a new bio-based plastic to replace conventional plastic.
KAIST (represented by President Kwang Hyung Lee) announced on the 20th of March that a research team led by Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering has developed microbial strains through systems metabolic engineering to produce various eco-friendly, bio-based poly(ester amide)s. The team collaborated with researchers from the Korea Research Institute of Chemical Technology (KRICT, President Young-Kook Lee) to analyze and confirm the properties of the resulting plastic.
Professor Sang Yup Lee’s research team designed new metabolic pathways that do not naturally exist in microorganisms, and developed a platform microbial strain capable of producing nine different types of poly(ester amide)s, including poly(3-hydroxybutyrate-ran-3-aminopropionate) and poly(3-hydroxybutyrate-ran-4-aminobutyrate).
Using glucose derived from abundant biomass sources such as waste wood and weeds, the team successfully produced poly(ester amide)s in an eco-friendly manner. The researchers also confirmed the potential for industrial-scale production by demonstrating high production efficiency (54.57 g/L) using fed-batch fermentation of the engineered strain.
In collaboration with researchers Haemin Jeong and Jihoon Shin from KRICT, the KAIST team analyzed the properties of the bio-based plastic and found that it exhibited characteristics similar to high-density polyethylene (HDPE). This means the new plastic is not only eco-friendly but also strong and durable enough to replace conventional plastics.
The engineered strains and strategies developed in this study are expected to be useful not only for producing various poly(ester amide)s but also for constructing metabolic pathways for the biosynthesis of other types of polymers.
Professor Sang Yup Lee stated, “This study is the first to demonstrate the possibility of producing poly(ester amide)s (plastics) through a renewable bio-based chemical process rather than relying on the petroleum-based chemical industry. We plan to further enhance the production yield and efficiency through continued research.”
The study was published online on March 17 in the international journal Nature Chemical Biology.
·Title: Biosynthesis of poly(ester amide)s in engineered Escherichia coli
·DOI: 10.1038/s41589-025-01842-2
·Authors: A total of seven authors including Tong Un Chae (KAIST, first author), So Young Choi (KAIST, second author), Da-Hee Ahn (KAIST, third author), Woo Dae Jang (KAIST, fourth author), Haemin Jeong (KRICT, fifth author), Jihoon Shin (KRICT, sixth author), and Sang Yup Lee (KAIST, corresponding author).
This research was supported by the Ministry of Science and ICT (MSIT) under the Eco-Friendly Chemical Technology Development Project as part of the "Next-Generation Biorefinery Technology Development to Lead the Bio-Chemical Industry" initiative (project led by Distinguished Professor Sang Yup Lee at KAIST).
2025.03.24 View 3694 -
 No More Touch Issues on Rainy Days! KAIST Develops Human-Like Tactile Sensor
Recent advancements in robotics have enabled machines to handle delicate objects like eggs with precision, thanks to highly integrated pressure sensors that provide detailed tactile feedback. However, even the most advanced robots struggle to accurately detect pressure in complex environments involving water, bending, or electromagnetic interference. A research team at KAIST has successfully developed a pressure sensor that operates stably without external interference, even on wet surfaces like a smartphone screen covered in water, achieving human-level tactile sensitivity.
KAIST (represented by President Kwang Hyung Lee) announced on the 10th of March that a research team led by Professor Jun-Bo Yoon from the School of Electrical Engineering has developed a high-resolution pressure sensor that remains unaffected by external interference such as "ghost touches" caused by moisture on touchscreens.
Capacitive pressure sensors, widely used in touch systems due to their simple structure and durability, are essential components of human-machine interface (HMI) technologies in smartphones, wearable devices, and robots. However, they are prone to malfunctions caused by water droplets, electromagnetic interference, and curves.
To address these issues, the research team investigated the root causes of interference in capacitive pressure sensors. They identified that the "fringe field" generated at the sensor’s edges is particularly susceptible to external disturbances.
The researchers concluded that, to fundamentally resolve this issue, suppressing the fringe field was necessary. Through theoretical analysis, they determined that reducing the electrode spacing to the nanometer scale could effectively minimize the fringe field to below a few percent.
Utilizing proprietary micro/nanofabrication techniques, the team developed a nanogap pressure sensor with an electrode spacing of 900 nanometers (nm). This newly developed sensor reliably detected pressure regardless of the material exerting force and remained unaffected by bending or electromagnetic interference.
Furthermore, the team successfully implemented an artificial tactile system utilizing the developed sensor’s characteristics. Human skin contains specialized pressure receptors called Merkel’s disks. To artificially mimic them, the exclusive detection of pressure was necessary, but hadn’t been achieved by conventional sensors.
Professor Yoon’s research team overcame these challenges, developing a sensor achieving a density comparable to Merkel’s discs and enabling wireless, high-precision pressure sensing.
To explore potential applications, the researcher also developed a force touch pad system, demonstrating its ability to capture pressure magnitude and distribution with high resolution and without interference.
Professor Yoon stated, “Our nanogap pressure sensor operates reliably even in rainy conditions or sweaty environments, eliminating common touch malfunctions. We believe this innovation will significantly enhance everyday user experiences.”
He added, “This technology has the potential to revolutionize various fields, including precision tactile sensors for robotics, medical wearable devices, and next-generation augmented reality (AR) and virtual reality (VR) interfaces.”
The study was led by Jae-Soon Yang (Ph.D.), Myung-Kun Chung (Ph.D. candidate), and Jae-Young Yoo (Assistant Professor at Sungkyunkwan University, a KAIST Ph.D. graduate). The research findings were published in Nature Communications on February 27, 2025. (Paper title: “Interference-Free Nanogap Pressure Sensor Array with High Spatial Resolution for Wireless Human-Machine Interface Applications”, DOI: 10.1038/s41467-025-57232-8)
This study was supported by the National Research Foundation of Korea’s Mid-Career Researcher Program and Leading Research Center Support Program.
2025.03.14 View 2520
No More Touch Issues on Rainy Days! KAIST Develops Human-Like Tactile Sensor
Recent advancements in robotics have enabled machines to handle delicate objects like eggs with precision, thanks to highly integrated pressure sensors that provide detailed tactile feedback. However, even the most advanced robots struggle to accurately detect pressure in complex environments involving water, bending, or electromagnetic interference. A research team at KAIST has successfully developed a pressure sensor that operates stably without external interference, even on wet surfaces like a smartphone screen covered in water, achieving human-level tactile sensitivity.
KAIST (represented by President Kwang Hyung Lee) announced on the 10th of March that a research team led by Professor Jun-Bo Yoon from the School of Electrical Engineering has developed a high-resolution pressure sensor that remains unaffected by external interference such as "ghost touches" caused by moisture on touchscreens.
Capacitive pressure sensors, widely used in touch systems due to their simple structure and durability, are essential components of human-machine interface (HMI) technologies in smartphones, wearable devices, and robots. However, they are prone to malfunctions caused by water droplets, electromagnetic interference, and curves.
To address these issues, the research team investigated the root causes of interference in capacitive pressure sensors. They identified that the "fringe field" generated at the sensor’s edges is particularly susceptible to external disturbances.
The researchers concluded that, to fundamentally resolve this issue, suppressing the fringe field was necessary. Through theoretical analysis, they determined that reducing the electrode spacing to the nanometer scale could effectively minimize the fringe field to below a few percent.
Utilizing proprietary micro/nanofabrication techniques, the team developed a nanogap pressure sensor with an electrode spacing of 900 nanometers (nm). This newly developed sensor reliably detected pressure regardless of the material exerting force and remained unaffected by bending or electromagnetic interference.
Furthermore, the team successfully implemented an artificial tactile system utilizing the developed sensor’s characteristics. Human skin contains specialized pressure receptors called Merkel’s disks. To artificially mimic them, the exclusive detection of pressure was necessary, but hadn’t been achieved by conventional sensors.
Professor Yoon’s research team overcame these challenges, developing a sensor achieving a density comparable to Merkel’s discs and enabling wireless, high-precision pressure sensing.
To explore potential applications, the researcher also developed a force touch pad system, demonstrating its ability to capture pressure magnitude and distribution with high resolution and without interference.
Professor Yoon stated, “Our nanogap pressure sensor operates reliably even in rainy conditions or sweaty environments, eliminating common touch malfunctions. We believe this innovation will significantly enhance everyday user experiences.”
He added, “This technology has the potential to revolutionize various fields, including precision tactile sensors for robotics, medical wearable devices, and next-generation augmented reality (AR) and virtual reality (VR) interfaces.”
The study was led by Jae-Soon Yang (Ph.D.), Myung-Kun Chung (Ph.D. candidate), and Jae-Young Yoo (Assistant Professor at Sungkyunkwan University, a KAIST Ph.D. graduate). The research findings were published in Nature Communications on February 27, 2025. (Paper title: “Interference-Free Nanogap Pressure Sensor Array with High Spatial Resolution for Wireless Human-Machine Interface Applications”, DOI: 10.1038/s41467-025-57232-8)
This study was supported by the National Research Foundation of Korea’s Mid-Career Researcher Program and Leading Research Center Support Program.
2025.03.14 View 2520 -
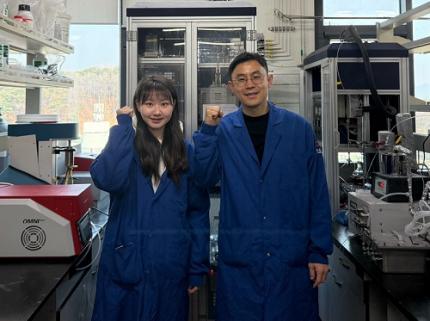 KAIST Develops World-Leading Ammonia Catalyst for Hydrogen Economy
Hydrogen production using renewable energy is a key technology for eco-friendly energy and chemical production. However, storing and transporting hydrogen remains a challenge. To address this, researchers worldwide are investigating methods to store hydrogen in the form of ammonia (NH₃), which is carbon-free and easier to liquify. A research team at KAIST has successfully developed a high-performance catalyst that enables ammonia synthesis at very low temperatures and pressures without energy loss.
KAIST (represented by President Kwang Hyung Lee) announced on the 11th of March that a research team led by Professor Minkee Choi from the Department of Chemical and Biomolecular Engineering has developed an innovative catalytic system that significantly enhances ammonia production while drastically reducing energy consumption and CO₂ emissions.
< (From left) Baek Ye-jun, Ph.D. candidate in the Department of Biochemical Engineering, Professor Choi Min-ki >
Currently, ammonia is produced using the Haber-Bosch process, a technology over a century old that relies on iron (Fe)-based catalysts. This method requires extreme conditions—temperatures above 500°C and pressures exceeding 100 atmospheres—resulting in enormous energy consumption and contributing significantly to global CO₂ emissions. Additionally, ammonia is primarily produced in large-scale industrial plants, leading to high distribution costs.
As an alternative, there is growing interest in an eco-friendly process that synthesizes ammonia using green hydrogen—produced via water electrolysis—under mild conditions (300°C, 10 atmospheres). However, developing catalysts that can achieve high ammonia productivity at such low temperatures and pressures is essential, as current technologies struggle to maintain efficiency under these conditions.
The research team developed a novel catalyst by incorporating ruthenium (Ru) nanoparticles and highly basic barium oxide (BaO) particles onto a conductive carbon surface, allowing it to function like a chemical capacitor*.
*Capacitor: A device that stores electrical energy by separating positive and negative charges.
During ammonia synthesis, hydrogen molecules (H₂) first dissociate into hydrogen atoms (H) on the ruthenium catalyst. These hydrogen atoms are further split into protons (H⁺) and electrons (e⁻). The study revealed that the acidic protons are stored in the strongly basic BaO, while the remaining electrons are separated and stored in ruthenium and carbon.
This unique chemical capacitor effect significantly enhances the ruthenium catalyst's electron density, accelerating nitrogen (N₂) dissociation—the rate-limiting step of ammonia synthesis—thereby dramatically increasing catalytic activity.
Furthermore, the team discovered that optimizing the nanostructure of the carbon material further boosts the electron density of ruthenium, maximizing catalytic performance. As a result, the new catalyst demonstrated over seven times higher ammonia synthesis performance compared to state-of-the-art catalysts under mild conditions (300°C, 10 atm).
< Schematic diagram showing the mechanism of ruthenium catalyst activity enhancement by barium oxide cocatalyst >
Professor Minkee Choi stated, “This research has garnered significant attention for demonstrating that catalytic activity can be greatly enhanced by controlling electron transfer within a thermal catalytic reaction system, not just in electrochemical processes.”
He further explained, “Our findings confirm that high-performance catalysts can enable efficient ammonia synthesis under low-temperature and low-pressure conditions. This could shift ammonia production from centralized, large-scale industrial plants to decentralized, small-scale production, making the hydrogen economy more sustainable and flexible.”
The study was led by Professor Minkee Choi as corresponding author and Yaejun Baik, a Ph.D. candidate, as first author. The research findings were published in Nature Catalysis on February 24.
(Paper title: “Electron and proton storage on separate Ru and BaO domains mediated by conductive low-work-function carbon to accelerate ammonia synthesis,” https://doi.org/10.1038/s41929-025-01302-z)
This research was supported by the Korea Institute of Energy Research and the National Research Foundation of Korea.
2025.03.11 View 2202
KAIST Develops World-Leading Ammonia Catalyst for Hydrogen Economy
Hydrogen production using renewable energy is a key technology for eco-friendly energy and chemical production. However, storing and transporting hydrogen remains a challenge. To address this, researchers worldwide are investigating methods to store hydrogen in the form of ammonia (NH₃), which is carbon-free and easier to liquify. A research team at KAIST has successfully developed a high-performance catalyst that enables ammonia synthesis at very low temperatures and pressures without energy loss.
KAIST (represented by President Kwang Hyung Lee) announced on the 11th of March that a research team led by Professor Minkee Choi from the Department of Chemical and Biomolecular Engineering has developed an innovative catalytic system that significantly enhances ammonia production while drastically reducing energy consumption and CO₂ emissions.
< (From left) Baek Ye-jun, Ph.D. candidate in the Department of Biochemical Engineering, Professor Choi Min-ki >
Currently, ammonia is produced using the Haber-Bosch process, a technology over a century old that relies on iron (Fe)-based catalysts. This method requires extreme conditions—temperatures above 500°C and pressures exceeding 100 atmospheres—resulting in enormous energy consumption and contributing significantly to global CO₂ emissions. Additionally, ammonia is primarily produced in large-scale industrial plants, leading to high distribution costs.
As an alternative, there is growing interest in an eco-friendly process that synthesizes ammonia using green hydrogen—produced via water electrolysis—under mild conditions (300°C, 10 atmospheres). However, developing catalysts that can achieve high ammonia productivity at such low temperatures and pressures is essential, as current technologies struggle to maintain efficiency under these conditions.
The research team developed a novel catalyst by incorporating ruthenium (Ru) nanoparticles and highly basic barium oxide (BaO) particles onto a conductive carbon surface, allowing it to function like a chemical capacitor*.
*Capacitor: A device that stores electrical energy by separating positive and negative charges.
During ammonia synthesis, hydrogen molecules (H₂) first dissociate into hydrogen atoms (H) on the ruthenium catalyst. These hydrogen atoms are further split into protons (H⁺) and electrons (e⁻). The study revealed that the acidic protons are stored in the strongly basic BaO, while the remaining electrons are separated and stored in ruthenium and carbon.
This unique chemical capacitor effect significantly enhances the ruthenium catalyst's electron density, accelerating nitrogen (N₂) dissociation—the rate-limiting step of ammonia synthesis—thereby dramatically increasing catalytic activity.
Furthermore, the team discovered that optimizing the nanostructure of the carbon material further boosts the electron density of ruthenium, maximizing catalytic performance. As a result, the new catalyst demonstrated over seven times higher ammonia synthesis performance compared to state-of-the-art catalysts under mild conditions (300°C, 10 atm).
< Schematic diagram showing the mechanism of ruthenium catalyst activity enhancement by barium oxide cocatalyst >
Professor Minkee Choi stated, “This research has garnered significant attention for demonstrating that catalytic activity can be greatly enhanced by controlling electron transfer within a thermal catalytic reaction system, not just in electrochemical processes.”
He further explained, “Our findings confirm that high-performance catalysts can enable efficient ammonia synthesis under low-temperature and low-pressure conditions. This could shift ammonia production from centralized, large-scale industrial plants to decentralized, small-scale production, making the hydrogen economy more sustainable and flexible.”
The study was led by Professor Minkee Choi as corresponding author and Yaejun Baik, a Ph.D. candidate, as first author. The research findings were published in Nature Catalysis on February 24.
(Paper title: “Electron and proton storage on separate Ru and BaO domains mediated by conductive low-work-function carbon to accelerate ammonia synthesis,” https://doi.org/10.1038/s41929-025-01302-z)
This research was supported by the Korea Institute of Energy Research and the National Research Foundation of Korea.
2025.03.11 View 2202 -
 KAIST achieves quantum entanglement essential for quantum error correction
Quantum computing is a technology capable of solving complex problems that classical computers struggle with. To perform accurate computations, quantum computers must correct errors that arise during operations. However, generating the quantum entanglement necessary for quantum error correction has long been considered a major challenge.
< Photo 1. (From left) Students Young-Do Yoon and Chan Roh of the Master's and Doctoral Integrated Program of the Department of Physics poses with Professor Young-Sik Ra and Student Geunhee Gwak of the same program >
KAIST (represented by President Kwang Hyung Lee) announced on the 25th of February that a research team led by Professor Young-Sik Ra from the Department of Physics has successfully implemented a three-dimensional cluster quantum entangled state, a key component for quantum error correction, through experimental demonstration.
Measurement-based quantum computing is an emerging paradigm that implements quantum computations by measuring specially entangled cluster states. The core of this approach lies in the generation of these cluster quantum entangled states, with two-dimensional cluster states commonly used for universal quantum computing.
However, to advance towards fault-tolerant quantum computing, which can correct quantum errors occurring during computations, a more complex three-dimensional cluster state is required. While previous studies have reported the generation of two-dimensional cluster states, experimental implementation of the three-dimensional cluster states necessary for fault-tolerant quantum computing had remained elusive due to the extreme complexity of their entanglement structure.
< Figure 1. (a) Experimental schematic. A pulse laser with a wavelength of 800 nm is converted into a pulse laser with a wavelength of 400 nm through second harmonic generation, and this is incident on a nonlinear crystal (PPKTP) to generate multiple quantum entanglement sources. (b) Generation of a 3D cluster state through optical mode basis change >
The research team overcame this challenge by developing a technique to control femtosecond time-frequency modes, successfully generating a three-dimensional cluster quantum entangled state for the first time.
The team directed a femtosecond laser into a nonlinear crystal, simultaneously generating quantum light sources across multiple frequency modes. (A femtosecond laser is a device that emits ultrashort, high-intensity light pulses.) Using this approach, they successfully created a three-dimensional cluster quantum entangled state.
Professor Young-Sik Ra noted, “This study marks the first successful demonstration of a three-dimensional cluster quantum entangled state, which was previously difficult to achieve with existing technology. This breakthrough is expected to serve as a crucial stepping stone for future research in measurement-based and fault-tolerant quantum computing.”
< Figure 2. Results of 3D cluster state generation. (a) Nullifier measurement of the cluster state. (b) 3D cluster state reconstructed using quantum state tomography. (c) Confirmation of quantum entanglement characteristics of the 3D cluster state >
The study was published online in Nature Photonics on February 24, 2025. The first author is Chan Roh, a Ph.D. candidate in KAIST’s integrated master’s and doctoral program, with Geunhee Gwak and Youngdo Yoon contributing as co-authors. (Paper title: “Generation of Three-Dimensional Cluster Entangled State”, DOI: 10.1038/s41566-025-01631-2)
This research was supported by the National Research Foundation of Korea (Quantum Computing Technology Development Program, Mid-Career Researcher Support Program, and Quantum Simulator for Materials Innovation Program), the Institute for Information & Communications Technology Planning & Evaluation (Quantum Internet Core Technology Program, University ICT Research Center Support Program), and the U.S. Air Force Research Laboratory.
2025.02.25 View 2207
KAIST achieves quantum entanglement essential for quantum error correction
Quantum computing is a technology capable of solving complex problems that classical computers struggle with. To perform accurate computations, quantum computers must correct errors that arise during operations. However, generating the quantum entanglement necessary for quantum error correction has long been considered a major challenge.
< Photo 1. (From left) Students Young-Do Yoon and Chan Roh of the Master's and Doctoral Integrated Program of the Department of Physics poses with Professor Young-Sik Ra and Student Geunhee Gwak of the same program >
KAIST (represented by President Kwang Hyung Lee) announced on the 25th of February that a research team led by Professor Young-Sik Ra from the Department of Physics has successfully implemented a three-dimensional cluster quantum entangled state, a key component for quantum error correction, through experimental demonstration.
Measurement-based quantum computing is an emerging paradigm that implements quantum computations by measuring specially entangled cluster states. The core of this approach lies in the generation of these cluster quantum entangled states, with two-dimensional cluster states commonly used for universal quantum computing.
However, to advance towards fault-tolerant quantum computing, which can correct quantum errors occurring during computations, a more complex three-dimensional cluster state is required. While previous studies have reported the generation of two-dimensional cluster states, experimental implementation of the three-dimensional cluster states necessary for fault-tolerant quantum computing had remained elusive due to the extreme complexity of their entanglement structure.
< Figure 1. (a) Experimental schematic. A pulse laser with a wavelength of 800 nm is converted into a pulse laser with a wavelength of 400 nm through second harmonic generation, and this is incident on a nonlinear crystal (PPKTP) to generate multiple quantum entanglement sources. (b) Generation of a 3D cluster state through optical mode basis change >
The research team overcame this challenge by developing a technique to control femtosecond time-frequency modes, successfully generating a three-dimensional cluster quantum entangled state for the first time.
The team directed a femtosecond laser into a nonlinear crystal, simultaneously generating quantum light sources across multiple frequency modes. (A femtosecond laser is a device that emits ultrashort, high-intensity light pulses.) Using this approach, they successfully created a three-dimensional cluster quantum entangled state.
Professor Young-Sik Ra noted, “This study marks the first successful demonstration of a three-dimensional cluster quantum entangled state, which was previously difficult to achieve with existing technology. This breakthrough is expected to serve as a crucial stepping stone for future research in measurement-based and fault-tolerant quantum computing.”
< Figure 2. Results of 3D cluster state generation. (a) Nullifier measurement of the cluster state. (b) 3D cluster state reconstructed using quantum state tomography. (c) Confirmation of quantum entanglement characteristics of the 3D cluster state >
The study was published online in Nature Photonics on February 24, 2025. The first author is Chan Roh, a Ph.D. candidate in KAIST’s integrated master’s and doctoral program, with Geunhee Gwak and Youngdo Yoon contributing as co-authors. (Paper title: “Generation of Three-Dimensional Cluster Entangled State”, DOI: 10.1038/s41566-025-01631-2)
This research was supported by the National Research Foundation of Korea (Quantum Computing Technology Development Program, Mid-Career Researcher Support Program, and Quantum Simulator for Materials Innovation Program), the Institute for Information & Communications Technology Planning & Evaluation (Quantum Internet Core Technology Program, University ICT Research Center Support Program), and the U.S. Air Force Research Laboratory.
2025.02.25 View 2207 -
 KAIST Research Team Develops an AI Framework Capable of Overcoming the Strength-Ductility Dilemma in Additive-manufactured Titanium Alloys
<(From Left) Ph.D. Student Jaejung Park and Professor Seungchul Lee of KAIST Department of Mechanical Engineering and , Professor Hyoung Seop Kim of POSTECH, and M.S.–Ph.D. Integrated Program Student Jeong Ah Lee of POSTECH. >
The KAIST research team led by Professor Seungchul Lee from Department of Mechanical Engineering, in collaboration with Professor Hyoung Seop Kim’s team at POSTECH, successfully overcame the strength–ductility dilemma of Ti 6Al 4V alloy using artificial intelligence, enabling the production of high strength, high ductility metal products. The AI developed by the team accurately predicts mechanical properties based on various 3D printing process parameters while also providing uncertainty information, and it uses both to recommend process parameters that hold high promise for 3D printing.
Among various 3D printing technologies, laser powder bed fusion is an innovative method for manufacturing Ti-6Al-4V alloy, renowned for its high strength and bio-compatibility. However, this alloy made via 3D printing has traditionally faced challenges in simultaneously achieving high strength and high ductility. Although there have been attempts to address this issue by adjusting both the printing process parameters and heat treatment conditions, the vast number of possible combinations made it difficult to explore them all through experiments and simulations alone.
The active learning framework developed by the team quickly explores a wide range of 3D printing process parameters and heat treatment conditions to recommend those expected to improve both strength and ductility of the alloy. These recommendations are based on the AI model’s predictions of ultimate tensile strength and total elongation along with associated uncertainty information for each set of process parameters and heat treatment conditions. The recommended conditions are then validated by performing 3D printing and tensile tests to obtain the true mechanical property values. These new data are incorporated into further AI model training, and through iterative exploration, the optimal process parameters and heat treatment conditions for producing high-performance alloys were determined in only five iterations. With these optimized conditions, the 3D printed Ti-6Al-4V alloy achieved an ultimate tensile strength of 1190 MPa and a total elongation of 16.5%, successfully overcoming the strength–ductility dilemma.
Professor Seungchul Lee commented, “In this study, by optimizing the 3D printing process parameters and heat treatment conditions, we were able to develop a high-strength, high-ductility Ti-6Al-4V alloy with minimal experimentation trials. Compared to previous studies, we produced an alloy with a similar ultimate tensile strength but higher total elongation, as well as that with a similar elongation but greater ultimate tensile strength.” He added, “Furthermore, if our approach is applied not only to mechanical properties but also to other properties such as thermal conductivity and thermal expansion, we anticipate that it will enable efficient exploration of 3D printing process parameters and heat treatment conditions.”
This study was published in Nature Communications on January 22 (https://doi.org/10.1038/s41467-025-56267-1), and the research was supported by the National Research Foundation of Korea’s Nano & Material Technology Development Program and the Leading Research Center Program.
2025.02.21 View 3733
KAIST Research Team Develops an AI Framework Capable of Overcoming the Strength-Ductility Dilemma in Additive-manufactured Titanium Alloys
<(From Left) Ph.D. Student Jaejung Park and Professor Seungchul Lee of KAIST Department of Mechanical Engineering and , Professor Hyoung Seop Kim of POSTECH, and M.S.–Ph.D. Integrated Program Student Jeong Ah Lee of POSTECH. >
The KAIST research team led by Professor Seungchul Lee from Department of Mechanical Engineering, in collaboration with Professor Hyoung Seop Kim’s team at POSTECH, successfully overcame the strength–ductility dilemma of Ti 6Al 4V alloy using artificial intelligence, enabling the production of high strength, high ductility metal products. The AI developed by the team accurately predicts mechanical properties based on various 3D printing process parameters while also providing uncertainty information, and it uses both to recommend process parameters that hold high promise for 3D printing.
Among various 3D printing technologies, laser powder bed fusion is an innovative method for manufacturing Ti-6Al-4V alloy, renowned for its high strength and bio-compatibility. However, this alloy made via 3D printing has traditionally faced challenges in simultaneously achieving high strength and high ductility. Although there have been attempts to address this issue by adjusting both the printing process parameters and heat treatment conditions, the vast number of possible combinations made it difficult to explore them all through experiments and simulations alone.
The active learning framework developed by the team quickly explores a wide range of 3D printing process parameters and heat treatment conditions to recommend those expected to improve both strength and ductility of the alloy. These recommendations are based on the AI model’s predictions of ultimate tensile strength and total elongation along with associated uncertainty information for each set of process parameters and heat treatment conditions. The recommended conditions are then validated by performing 3D printing and tensile tests to obtain the true mechanical property values. These new data are incorporated into further AI model training, and through iterative exploration, the optimal process parameters and heat treatment conditions for producing high-performance alloys were determined in only five iterations. With these optimized conditions, the 3D printed Ti-6Al-4V alloy achieved an ultimate tensile strength of 1190 MPa and a total elongation of 16.5%, successfully overcoming the strength–ductility dilemma.
Professor Seungchul Lee commented, “In this study, by optimizing the 3D printing process parameters and heat treatment conditions, we were able to develop a high-strength, high-ductility Ti-6Al-4V alloy with minimal experimentation trials. Compared to previous studies, we produced an alloy with a similar ultimate tensile strength but higher total elongation, as well as that with a similar elongation but greater ultimate tensile strength.” He added, “Furthermore, if our approach is applied not only to mechanical properties but also to other properties such as thermal conductivity and thermal expansion, we anticipate that it will enable efficient exploration of 3D printing process parameters and heat treatment conditions.”
This study was published in Nature Communications on January 22 (https://doi.org/10.1038/s41467-025-56267-1), and the research was supported by the National Research Foundation of Korea’s Nano & Material Technology Development Program and the Leading Research Center Program.
2025.02.21 View 3733 -
 KAIST Develops Stretchable Displays Featuring 25% Expansion Without Image Distortion
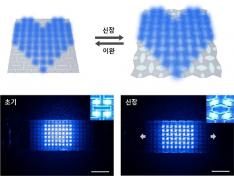
Stretchable displays, praised for their spatial efficiency, design flexibility, and human-like flexibility, are seen as the next generation of display technology. A team of Korean researchers has developed a stretchable display that can expand by 25% while maintaining clear image quality without distortion. It can also stretch and contract up to 5,000 times at 15% expansion without any performance degradation, making it the first deformation-free stretchable display with a negative Poisson's ratio* developed in Korea.
*Poisson’s ratio of -1: A ratio where both width and length stretch equally, expressed as a negative value. A positive Poisson's ratio represents the ratio where horizontal stretching leads to vertical contraction, which is the case for most materials.
KAIST (represented by President Kwang-Hyung Lee) announced on the 20th of August that a research team led by Professor Byeong-Soo Bae of the Department of Materials Science and Engineering (Director of the Wearable Platform Materials Technology Center) , in collaboration with the Korea Institute of Machinery & Materials (President Seoghyeon Ryu), successfully developed a stretchable display substrate that suppresses image distortion through omnidirectional stretchability.
Currently, most stretchable displays are made with highly elastic elastomer* materials, but these materials possess a positive Poisson's ratio, causing unavoidable image distortion when the display is stretched.
*Elastomer: A polymer with elasticity similar to rubber.
To address this, the introduction of auxetic* meta-structures has been gaining attention. Unlike conventional materials, auxetic structures have a unique 'negative Poisson's ratio,' expanding in all directions when stretched in just one direction. However, traditional auxetic structures contain many empty spaces, limiting their stability and usability in display substrates.
*Auxetic structure: A special geometric structure that exhibits a negative Poisson's ratio.
To tackle the issue of image distortion, Professor Bae's research team developed a method to create a seamless surface for the auxetic meta-structure, achieving the ideal negative Poisson's ratio of -1 and overcoming the biggest challenge in auxetic meta-structures.
To overcome the second issue of elastic modulus*, the team inserted a textile made of glass fiber bundles with a diameter of just 25 micrometers (a quarter of the thickness of human hair) into the elastomer material. They then filled the empty spaces with the same elastomer, creating a flat and stable integrated film without gaps.
*Elastic Modulus: The ratio that indicates the extent of deformation when force is applied to a material. A higher elastic modulus means that the material is less likely to deform under force.
The research team theoretically identified that the difference in elasticity between the auxetic structure and the elastomer material directly influences the negative Poisson's ratio and successfully achieved an elasticity difference of over 230,000 times, producing a film with a Poisson's ratio of -1, the theoretical limit.
< Figure 2. Deformation of S-AUX film. a) Configurations and visualized principal strain distribution of the optimized S-AUX film at various strain rates. b) Biaxial stretching image. While pristine elastomer shrinks in the directions that were not stretched, S-AUX film developed in this study expands in all directions simultaneously while maintaining its original shape. >
Professor Byeong-Soo Bae, who led the study, explained, "Preventing image distortion using auxetic structures in stretchable displays is a core technology, but it has faced challenges due to the many empty spaces in the surface, making it difficult to use as a substrate. This research outcome is expected to significantly accelerate commercialization through high-resolution, distortion-free stretchable display applications that utilize the entire surface."
This study, co-authored by Dr. Yung Lee from KAIST’s Department of Materials Science and Engineering and Dr. Bongkyun Jang from the Korea Institute of Machinery & Materials, was published on August 20th in the international journal Nature Communications under the title "A seamless auxetic substrate with a negative Poisson's ratio of –1".
The research was supported by the Wearable Platform Materials Technology Center at KAIST, the Korea Institute of Machinery & Materials, and LG Display.
< Figure 3. Structural configuration of the distortion-free display components on the S-AUX film and a contour image of a micro-LED chip transferred onto the S-AUX film. >
< Figure 4. Schematic illustrations and photographic images of the S-AUX film-based image: distortion-free display in its stretched state and released state. >
2024.09.20 View 5070
KAIST Develops Stretchable Displays Featuring 25% Expansion Without Image Distortion
Stretchable displays, praised for their spatial efficiency, design flexibility, and human-like flexibility, are seen as the next generation of display technology. A team of Korean researchers has developed a stretchable display that can expand by 25% while maintaining clear image quality without distortion. It can also stretch and contract up to 5,000 times at 15% expansion without any performance degradation, making it the first deformation-free stretchable display with a negative Poisson's ratio* developed in Korea.
*Poisson’s ratio of -1: A ratio where both width and length stretch equally, expressed as a negative value. A positive Poisson's ratio represents the ratio where horizontal stretching leads to vertical contraction, which is the case for most materials.
KAIST (represented by President Kwang-Hyung Lee) announced on the 20th of August that a research team led by Professor Byeong-Soo Bae of the Department of Materials Science and Engineering (Director of the Wearable Platform Materials Technology Center) , in collaboration with the Korea Institute of Machinery & Materials (President Seoghyeon Ryu), successfully developed a stretchable display substrate that suppresses image distortion through omnidirectional stretchability.
Currently, most stretchable displays are made with highly elastic elastomer* materials, but these materials possess a positive Poisson's ratio, causing unavoidable image distortion when the display is stretched.
*Elastomer: A polymer with elasticity similar to rubber.
To address this, the introduction of auxetic* meta-structures has been gaining attention. Unlike conventional materials, auxetic structures have a unique 'negative Poisson's ratio,' expanding in all directions when stretched in just one direction. However, traditional auxetic structures contain many empty spaces, limiting their stability and usability in display substrates.
*Auxetic structure: A special geometric structure that exhibits a negative Poisson's ratio.
To tackle the issue of image distortion, Professor Bae's research team developed a method to create a seamless surface for the auxetic meta-structure, achieving the ideal negative Poisson's ratio of -1 and overcoming the biggest challenge in auxetic meta-structures.
To overcome the second issue of elastic modulus*, the team inserted a textile made of glass fiber bundles with a diameter of just 25 micrometers (a quarter of the thickness of human hair) into the elastomer material. They then filled the empty spaces with the same elastomer, creating a flat and stable integrated film without gaps.
*Elastic Modulus: The ratio that indicates the extent of deformation when force is applied to a material. A higher elastic modulus means that the material is less likely to deform under force.
The research team theoretically identified that the difference in elasticity between the auxetic structure and the elastomer material directly influences the negative Poisson's ratio and successfully achieved an elasticity difference of over 230,000 times, producing a film with a Poisson's ratio of -1, the theoretical limit.
< Figure 2. Deformation of S-AUX film. a) Configurations and visualized principal strain distribution of the optimized S-AUX film at various strain rates. b) Biaxial stretching image. While pristine elastomer shrinks in the directions that were not stretched, S-AUX film developed in this study expands in all directions simultaneously while maintaining its original shape. >
Professor Byeong-Soo Bae, who led the study, explained, "Preventing image distortion using auxetic structures in stretchable displays is a core technology, but it has faced challenges due to the many empty spaces in the surface, making it difficult to use as a substrate. This research outcome is expected to significantly accelerate commercialization through high-resolution, distortion-free stretchable display applications that utilize the entire surface."
This study, co-authored by Dr. Yung Lee from KAIST’s Department of Materials Science and Engineering and Dr. Bongkyun Jang from the Korea Institute of Machinery & Materials, was published on August 20th in the international journal Nature Communications under the title "A seamless auxetic substrate with a negative Poisson's ratio of –1".
The research was supported by the Wearable Platform Materials Technology Center at KAIST, the Korea Institute of Machinery & Materials, and LG Display.
< Figure 3. Structural configuration of the distortion-free display components on the S-AUX film and a contour image of a micro-LED chip transferred onto the S-AUX film. >
< Figure 4. Schematic illustrations and photographic images of the S-AUX film-based image: distortion-free display in its stretched state and released state. >
2024.09.20 View 5070 -
 KAIST presents strategies for Holotomography in advanced bio research
Measuring and analyzing three-dimensional (3D) images of live cells and tissues is considered crucial in advanced fields of biology and medicine. Organoids, which are 3D structures that mimic organs, are particular examples that significantly benefits 3D live imaging. Organoids provide effective alternatives to animal testing in the drug development processes, and can rapidly determine personalized medicine. On the other hand, active researches are ongoing to utilize organoids for organ replacement.
< Figure 1. Schematic illustration of holotomography compared to X-ray CT. Similar to CT, they share the commonality of measuring the optical properties of an unlabeled specimen in three dimensions. Instead of X-rays, holotomography irradiates light in the visible range, and provides refractive index measurements of transparent specimens rather than absorptivity. While CT obtains three-dimensional information only through mechanical rotation of the irradiating light, holotomography can replace this by applying wavefront control technology in the visible range. >
Organelle-level observation of 3D biological specimens such as organoids and stem cell colonies without staining or preprocessing holds significant implications for both innovating basic research and bioindustrial applications related to regenerative medicine and bioindustrial applications.
Holotomography (HT) is a 3D optical microscopy that implements 3D reconstruction analogous to that of X-ray computed tomography (CT). Although HT and CT share a similar theoretical background, HT facilitates high-resolution examination inside cells and tissues, instead of the human body. HT obtains 3D images of cells and tissues at the organelle level without chemical or genetic labeling, thus overcomes various challenges of existing methods in bio research and industry. Its potential is highlighted in research fields where sample physiology must not be disrupted, such as regenerative medicine, personalized medicine, and infertility treatment.
< Figure 2. Label-free 3D imaging of diverse live cells. Time-lapse image of Hep3B cells illustrating subcellular morphology changes upon H2O2 treatment, followed by cellular recovery after returning to the regular cell culture medium. >
This paper introduces the advantages and broad applicability of HT to biomedical researchers, while presenting an overview of principles and future technical challenges to optical researchers. It showcases various cases of applying HT in studies such as 3D biology, regenerative medicine, and cancer research, as well as suggesting future optical development. Also, it categorizes HT based on the light source, to describe the principles, limitations, and improvements of each category in detail. Particularly, the paper addresses strategies for deepening cell and organoid studies by introducing artificial intelligence (AI) to HT.
Due to its potential to drive advanced bioindustry, HT is attracting interest and investment from universities and corporates worldwide. The KAIST research team has been leading this international field by developing core technologies and carrying out key application researches throughout the last decade.
< Figure 3. Various types of cells and organelles that make up the imaging barrier of a living intestinal organoid can be observed using holotomography. >
This paper, co-authored by Dr. Geon Kim from KAIST Research Center for Natural Sciences, Professor Ki-Jun Yoon's team from the Department of Biological Sciences, Director Bon-Kyoung Koo's team from the Institute for Basic Science (IBS) Center for Genome Engineering, and Dr. Seongsoo Lee's team from the Korea Basic Science Institute (KBSI), was published in 'Nature Reviews Methods Primers' on the 25th of July. This research was supported by the Leader Grant and Basic Science Research Program of the National Research Foundation, the Hologram Core Technology Development Grant of the Ministry of Science and ICT, the Nano and Material Technology Development Project, and the Health and Medical R&D Project of the Ministry of Health and Welfare.
2024.07.30 View 4759
KAIST presents strategies for Holotomography in advanced bio research
Measuring and analyzing three-dimensional (3D) images of live cells and tissues is considered crucial in advanced fields of biology and medicine. Organoids, which are 3D structures that mimic organs, are particular examples that significantly benefits 3D live imaging. Organoids provide effective alternatives to animal testing in the drug development processes, and can rapidly determine personalized medicine. On the other hand, active researches are ongoing to utilize organoids for organ replacement.
< Figure 1. Schematic illustration of holotomography compared to X-ray CT. Similar to CT, they share the commonality of measuring the optical properties of an unlabeled specimen in three dimensions. Instead of X-rays, holotomography irradiates light in the visible range, and provides refractive index measurements of transparent specimens rather than absorptivity. While CT obtains three-dimensional information only through mechanical rotation of the irradiating light, holotomography can replace this by applying wavefront control technology in the visible range. >
Organelle-level observation of 3D biological specimens such as organoids and stem cell colonies without staining or preprocessing holds significant implications for both innovating basic research and bioindustrial applications related to regenerative medicine and bioindustrial applications.
Holotomography (HT) is a 3D optical microscopy that implements 3D reconstruction analogous to that of X-ray computed tomography (CT). Although HT and CT share a similar theoretical background, HT facilitates high-resolution examination inside cells and tissues, instead of the human body. HT obtains 3D images of cells and tissues at the organelle level without chemical or genetic labeling, thus overcomes various challenges of existing methods in bio research and industry. Its potential is highlighted in research fields where sample physiology must not be disrupted, such as regenerative medicine, personalized medicine, and infertility treatment.
< Figure 2. Label-free 3D imaging of diverse live cells. Time-lapse image of Hep3B cells illustrating subcellular morphology changes upon H2O2 treatment, followed by cellular recovery after returning to the regular cell culture medium. >
This paper introduces the advantages and broad applicability of HT to biomedical researchers, while presenting an overview of principles and future technical challenges to optical researchers. It showcases various cases of applying HT in studies such as 3D biology, regenerative medicine, and cancer research, as well as suggesting future optical development. Also, it categorizes HT based on the light source, to describe the principles, limitations, and improvements of each category in detail. Particularly, the paper addresses strategies for deepening cell and organoid studies by introducing artificial intelligence (AI) to HT.
Due to its potential to drive advanced bioindustry, HT is attracting interest and investment from universities and corporates worldwide. The KAIST research team has been leading this international field by developing core technologies and carrying out key application researches throughout the last decade.
< Figure 3. Various types of cells and organelles that make up the imaging barrier of a living intestinal organoid can be observed using holotomography. >
This paper, co-authored by Dr. Geon Kim from KAIST Research Center for Natural Sciences, Professor Ki-Jun Yoon's team from the Department of Biological Sciences, Director Bon-Kyoung Koo's team from the Institute for Basic Science (IBS) Center for Genome Engineering, and Dr. Seongsoo Lee's team from the Korea Basic Science Institute (KBSI), was published in 'Nature Reviews Methods Primers' on the 25th of July. This research was supported by the Leader Grant and Basic Science Research Program of the National Research Foundation, the Hologram Core Technology Development Grant of the Ministry of Science and ICT, the Nano and Material Technology Development Project, and the Health and Medical R&D Project of the Ministry of Health and Welfare.
2024.07.30 View 4759 -
 Unraveling Mitochondrial DNA Mutations in Human Cells
Throughout our lifetime, cells accumulate DNA mutations, which contribute to genetic diversity, or “mosaicism”, among cells. These genomic mutations are pivotal for the aging process and the onset of various diseases, including cancer. Mitochondria, essential cellular organelles involved in energy metabolism and apoptosis, possess their own DNA, which are susceptible to mutations. However, studies on mtDNA mutations and mosaicism have been limited due to a variety of technical challenges.
Genomic scientists from KAIST have revealed the genetic mosaicism characterized by variations in mitochondrial DNA (mtDNA) across normal human cells. This study provides fundamental insights into understanding human aging and disease onset mechanisms.
The study, “Mitochondrial DNA mosaicism in normal human somatic cells,” was published in Nature Genetics on July 22. It was led by graduate student Jisong An under the supervision of Professor Young Seok Ju from the Graduate School of Medical Science and Engineering.
Researchers from Seoul National University College of Medicine, Yonsei University College of Medicine, Korea University College of Medicine, Washington University School of Medicine National Cancer Center, Seoul National University Hospital, Gangnam Severance Hospital and KAIST faculty startup company Inocras Inc. also participated in this study.
< Figure 1. a. Flow of experiment. b. Schematic diagram illustrating the origin and dynamics of mtDNA alterations across a lifetime. >
The study involved a bioinformatic analysis of whole-genome sequences from 2,096 single cells obtained from normal human colorectal epithelial tissue, fibroblasts, and blood collected from 31 individuals. The study highlights an average of three significant mtDNA differences between cells, with approximately 6% of these variations confirmed to be inherited as heteroplasmy from the mother.
Moreover, mutations significantly increased during tumorigenesis, with some mutations contributing to instability in mitochondrial RNA. Based on these findings, the study illustrates a computational model that comprehensively elucidates the evolution of mitochondria from embryonic development to aging and tumorigenesis.
This study systematically reveals the mechanisms behind mitochondrial DNA mosaicism in normal human cells, establishing a crucial foundation for understanding the impact of mtDNA on aging and disease onset.
Professor Ju remarked, “By systematically utilizing whole-genome big data, we can illuminate previously unknown phenomena in life sciences.” He emphasized the significance of the study, adding, “For the first time, we have established a method to systematically understand mitochondrial DNA changes occurring during human embryonic development, aging, and cancer development.”
This work was supported by the National Research Foundation of Korea and the Suh Kyungbae Foundation.
2024.07.24 View 4736
Unraveling Mitochondrial DNA Mutations in Human Cells
Throughout our lifetime, cells accumulate DNA mutations, which contribute to genetic diversity, or “mosaicism”, among cells. These genomic mutations are pivotal for the aging process and the onset of various diseases, including cancer. Mitochondria, essential cellular organelles involved in energy metabolism and apoptosis, possess their own DNA, which are susceptible to mutations. However, studies on mtDNA mutations and mosaicism have been limited due to a variety of technical challenges.
Genomic scientists from KAIST have revealed the genetic mosaicism characterized by variations in mitochondrial DNA (mtDNA) across normal human cells. This study provides fundamental insights into understanding human aging and disease onset mechanisms.
The study, “Mitochondrial DNA mosaicism in normal human somatic cells,” was published in Nature Genetics on July 22. It was led by graduate student Jisong An under the supervision of Professor Young Seok Ju from the Graduate School of Medical Science and Engineering.
Researchers from Seoul National University College of Medicine, Yonsei University College of Medicine, Korea University College of Medicine, Washington University School of Medicine National Cancer Center, Seoul National University Hospital, Gangnam Severance Hospital and KAIST faculty startup company Inocras Inc. also participated in this study.
< Figure 1. a. Flow of experiment. b. Schematic diagram illustrating the origin and dynamics of mtDNA alterations across a lifetime. >
The study involved a bioinformatic analysis of whole-genome sequences from 2,096 single cells obtained from normal human colorectal epithelial tissue, fibroblasts, and blood collected from 31 individuals. The study highlights an average of three significant mtDNA differences between cells, with approximately 6% of these variations confirmed to be inherited as heteroplasmy from the mother.
Moreover, mutations significantly increased during tumorigenesis, with some mutations contributing to instability in mitochondrial RNA. Based on these findings, the study illustrates a computational model that comprehensively elucidates the evolution of mitochondria from embryonic development to aging and tumorigenesis.
This study systematically reveals the mechanisms behind mitochondrial DNA mosaicism in normal human cells, establishing a crucial foundation for understanding the impact of mtDNA on aging and disease onset.
Professor Ju remarked, “By systematically utilizing whole-genome big data, we can illuminate previously unknown phenomena in life sciences.” He emphasized the significance of the study, adding, “For the first time, we have established a method to systematically understand mitochondrial DNA changes occurring during human embryonic development, aging, and cancer development.”
This work was supported by the National Research Foundation of Korea and the Suh Kyungbae Foundation.
2024.07.24 View 4736 -
 KAIST introduces microbial food as a strategy food production of the future
The global food crisis is increasing due to rapid population growth and declining food productivity to climate change. Moreover, today's food production and supply system emit a huge amount of carbon dioxide, reaching 30% of the total amount emitted by humanity, aggravating climate change. Sustainable and nutritious microbial food is attracting attention as a key to overcoming this impasse.
KAIST (President Kwang Hyung Lee) announced on April 12th that Research Professor Kyeong Rok Choi of the BioProcess Engineering Research Center and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering published a paper that proposes a direction of research on ‘microbial food production from sustainable raw materials.’
Microbial food refers to various foods and food ingredients produced using microorganisms. Microbial biomass contains a large amount of protein per unit in dry mass, comparable to that of meat, and emits the smallest amount of carbon dioxide and is required to produce a unit mass compared to various livestock, fish, shellfish, and crops. Since the amount of water and space requirement is small, it can be an eco-friendly, sustainable and highly nutritious food resource.
Fermented foods are the most readily available microbial foods around us. Although the proportion of microbial biomass in fermented foods is small, compounds with relatively low nutritional value, such as carbohydrates, are consumed during the fermentation process, and as microorganisms proliferate, the content of nutrients with higher nutritional value, such as proteins and vitamins, increases.
Various food compounds isolated and purified from biomass or culture media obtained through microbial culture are also a branch of microbial food. Examples that can be found around us include various amino acids, including monosodium glutamate, food proteins, enzymes, flavoring compounds, food colorings, and bioactive substances.
< Figure 1. Schematic diagram portraying various microbial biomass production strategies utlizing sustainable feedstocks >
Lastly, the most ultimate and fundamental form of microbial food can be said to be microbial biomass or extracts produced through microbial culture and foods cooked using them. A representative example is single-cell protein, which collectively refers to microbial biomass or microbial proteins extracted from it.
In this paper, the researchers comprehensively covered various non-edible raw materials and strategies for using them that can be used to produce microbial food in a more sustainable way. Furthermore, it covers various microbial foods that are actually produced in the industry using the relevant raw materials and their characteristics, as well as prospects for the production and generalization of sustainable microbial foods.
Research Professor Kyeong Rok Choi, the first author of this paper, said, “Microbial foods produced from various sustainable raw materials will soon be commonly encountered at our tables.” Second author Seok Yeong Jung, a doctoral student, also said, “Microbial foods of the future will not be limited foods consumed only out of a sense of obligation to the environment, but will be complete foods that are consumed by choice because of their nutritional value and taste.” In addition, Distinguished Professor Sang Yup Lee said, “It is time for the industry and academia, as well as the public and private sectors, to cooperate more closely so that more diverse microbial foods can be developed and supplied in order to create a sustainable society for ourselves and our descendants.”
< Figure 2. Compositions and environmental footprints of animal, plant and microbial biomass. >
This paper was published online on April 9 in ‘Nature Microbiology’ published by Nature.
※ Paper title: From sustainable feedstocks to microbial foods
※ Author information: Kyeong Rok Choi (first author), Seok Yeong Jung (second author) and Sang Yup Lee (corresponding author)
This research was conducted under the development of platform technologies of microbial cell factories for the next-generation biorefineries project (project leader KAIST Distinguished Professor Sang Yup Lee) supported by the Ministry of Science and ICT and the Cooperative Research Program for Agriculture Science and Technology Development (Project leader KAIST Research Professor Kyeong Rok Choi) of the Agricultural Microbiology Project Group (Director, Professor Pahn-Shick Chang) supported by the Rural Development Administration.
2024.04.12 View 7050
KAIST introduces microbial food as a strategy food production of the future
The global food crisis is increasing due to rapid population growth and declining food productivity to climate change. Moreover, today's food production and supply system emit a huge amount of carbon dioxide, reaching 30% of the total amount emitted by humanity, aggravating climate change. Sustainable and nutritious microbial food is attracting attention as a key to overcoming this impasse.
KAIST (President Kwang Hyung Lee) announced on April 12th that Research Professor Kyeong Rok Choi of the BioProcess Engineering Research Center and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering published a paper that proposes a direction of research on ‘microbial food production from sustainable raw materials.’
Microbial food refers to various foods and food ingredients produced using microorganisms. Microbial biomass contains a large amount of protein per unit in dry mass, comparable to that of meat, and emits the smallest amount of carbon dioxide and is required to produce a unit mass compared to various livestock, fish, shellfish, and crops. Since the amount of water and space requirement is small, it can be an eco-friendly, sustainable and highly nutritious food resource.
Fermented foods are the most readily available microbial foods around us. Although the proportion of microbial biomass in fermented foods is small, compounds with relatively low nutritional value, such as carbohydrates, are consumed during the fermentation process, and as microorganisms proliferate, the content of nutrients with higher nutritional value, such as proteins and vitamins, increases.
Various food compounds isolated and purified from biomass or culture media obtained through microbial culture are also a branch of microbial food. Examples that can be found around us include various amino acids, including monosodium glutamate, food proteins, enzymes, flavoring compounds, food colorings, and bioactive substances.
< Figure 1. Schematic diagram portraying various microbial biomass production strategies utlizing sustainable feedstocks >
Lastly, the most ultimate and fundamental form of microbial food can be said to be microbial biomass or extracts produced through microbial culture and foods cooked using them. A representative example is single-cell protein, which collectively refers to microbial biomass or microbial proteins extracted from it.
In this paper, the researchers comprehensively covered various non-edible raw materials and strategies for using them that can be used to produce microbial food in a more sustainable way. Furthermore, it covers various microbial foods that are actually produced in the industry using the relevant raw materials and their characteristics, as well as prospects for the production and generalization of sustainable microbial foods.
Research Professor Kyeong Rok Choi, the first author of this paper, said, “Microbial foods produced from various sustainable raw materials will soon be commonly encountered at our tables.” Second author Seok Yeong Jung, a doctoral student, also said, “Microbial foods of the future will not be limited foods consumed only out of a sense of obligation to the environment, but will be complete foods that are consumed by choice because of their nutritional value and taste.” In addition, Distinguished Professor Sang Yup Lee said, “It is time for the industry and academia, as well as the public and private sectors, to cooperate more closely so that more diverse microbial foods can be developed and supplied in order to create a sustainable society for ourselves and our descendants.”
< Figure 2. Compositions and environmental footprints of animal, plant and microbial biomass. >
This paper was published online on April 9 in ‘Nature Microbiology’ published by Nature.
※ Paper title: From sustainable feedstocks to microbial foods
※ Author information: Kyeong Rok Choi (first author), Seok Yeong Jung (second author) and Sang Yup Lee (corresponding author)
This research was conducted under the development of platform technologies of microbial cell factories for the next-generation biorefineries project (project leader KAIST Distinguished Professor Sang Yup Lee) supported by the Ministry of Science and ICT and the Cooperative Research Program for Agriculture Science and Technology Development (Project leader KAIST Research Professor Kyeong Rok Choi) of the Agricultural Microbiology Project Group (Director, Professor Pahn-Shick Chang) supported by the Rural Development Administration.
2024.04.12 View 7050