Kyungpook+National+University
-
 Mystery of Biological Plastic Synthesis Machinery Unveiled
Plastics and other polymers are used every day. These polymers are mostly made from fossil resources by refining petrochemicals. On the other hand, many microorganisms naturally synthesize polyesters known as polyhydroxyalkanoates (PHAs) as distinct granules inside cells.
PHAs are a family of microbial polyesters that have attracted much attention as biodegradable and biocompatible plastics and elastomers that can substitute petrochemical counterparts. There have been numerous papers and patents on gene cloning and metabolic engineering of PHA biosynthetic machineries, biochemical studies, and production of PHAs; simple Google search with “polyhydroxyalkanoates” yielded returns of 223,000 document pages. PHAs have always been considered amazing examples of biological polymer synthesis. It is astounding to see PHAs of 500 kDa to sometimes as high as 10,000 kDa can be synthesized in vivo by PHA synthase, the key polymerizing enzyme in PHA biosynthesis. They have attracted great interest in determining the crystal structure of PHA synthase over the last 30 years, but unfortunately without success. Thus, the characteristics and molecular mechanisms of PHA synthase were under a dark veil.
In two papers published back-to-back in Biotechnology Journal online on November 30, 2016, a Korean research team led by Professor Kyung-Jin Kim at Kyungpook National University and Distinguished Professor Sang Yup Lee at the Korea Advanced Institute of Science and Technology (KAIST) described the crystal structure of PHA synthase from Ralstonia eutropha, the best studied bacterium for PHA production, and reported the structural basis for the detailed molecular mechanisms of PHA biosynthesis. The crystal structure has been deposited to Protein Data Bank in February 2016. After deciphering the crystal structure of the catalytic domain of PHA synthase, in addition to other structural studies on whole enzyme and related proteins, the research team also performed experiments to elucidate the mechanisms of the enzyme reaction, validating detailed structures, enzyme engineering, and also N-terminal domain studies among others.
Through several biochemical studies based on crystal structure, the authors show that PHA synthase exists as a dimer and is divided into two distinct domains, the N-terminal domain (RePhaC1ND) and the C-terminal domain (RePhaC1CD). The RePhaC1CD catalyzes the polymerization reaction via a non-processive ping-pong mechanism using a Cys-His-Asp catalytic triad. The two catalytic sites of the RePhaC1CD dimer are positioned 33.4 Å apart, suggesting that the polymerization reaction occurs independently at each site. This study also presents the structure-based mechanisms for substrate specificities of various PHA synthases from different classes.
Professor Sang Yup Lee, who has worked on this topic for more than 20 years, said,
“The results and information presented in these two papers have long been awaited not only in the PHA community, but also metabolic engineering, bacteriology/microbiology, and in general biological sciences communities. The structural information on PHA synthase together with the recently deciphered reaction mechanisms will be valuable for understanding the detailed mechanisms of biosynthesizing this important energy/redox storage material, and also for the rational engineering of PHA synthases to produce designer bioplastics from various monomers more efficiently.”
Indeed, these two papers published in Biotechnology Journal finally reveal the 30-year mystery of machinery of biological polyester synthesis, and will serve as the essential compass in creating designer and more efficient bioplastic machineries.
References:
Jieun Kim, Yeo-Jin Kim, So Young Choi, Sang Yup Lee and Kyung-Jin Kim. “Crystal structure of Ralstonia eutropha polyhydroxyalkanoate synthase C-terminal domain and reaction mechanisms” Biotechnology Journal DOI: 10.1002/biot.201600648
http://onlinelibrary.wiley.com/doi/10.1002/biot.201600648/abstract
Yeo-Jin Kim, So Young Choi, Jieun Kim, Kyeong Sik Jin, Sang Yup Lee and Kyung-Jin Kim. “Structure and function of the N-terminal domain of Ralstonia eutropha polyhydroxyalkanoate synthase, and the proposed structure and mechanisms of the whole enzyme” Biotechnology Journal DOI: 10.1002/biot.201600649
http://onlinelibrary.wiley.com/doi/10.1002/biot.201600649/abstract
2016.12.02 View 12149
Mystery of Biological Plastic Synthesis Machinery Unveiled
Plastics and other polymers are used every day. These polymers are mostly made from fossil resources by refining petrochemicals. On the other hand, many microorganisms naturally synthesize polyesters known as polyhydroxyalkanoates (PHAs) as distinct granules inside cells.
PHAs are a family of microbial polyesters that have attracted much attention as biodegradable and biocompatible plastics and elastomers that can substitute petrochemical counterparts. There have been numerous papers and patents on gene cloning and metabolic engineering of PHA biosynthetic machineries, biochemical studies, and production of PHAs; simple Google search with “polyhydroxyalkanoates” yielded returns of 223,000 document pages. PHAs have always been considered amazing examples of biological polymer synthesis. It is astounding to see PHAs of 500 kDa to sometimes as high as 10,000 kDa can be synthesized in vivo by PHA synthase, the key polymerizing enzyme in PHA biosynthesis. They have attracted great interest in determining the crystal structure of PHA synthase over the last 30 years, but unfortunately without success. Thus, the characteristics and molecular mechanisms of PHA synthase were under a dark veil.
In two papers published back-to-back in Biotechnology Journal online on November 30, 2016, a Korean research team led by Professor Kyung-Jin Kim at Kyungpook National University and Distinguished Professor Sang Yup Lee at the Korea Advanced Institute of Science and Technology (KAIST) described the crystal structure of PHA synthase from Ralstonia eutropha, the best studied bacterium for PHA production, and reported the structural basis for the detailed molecular mechanisms of PHA biosynthesis. The crystal structure has been deposited to Protein Data Bank in February 2016. After deciphering the crystal structure of the catalytic domain of PHA synthase, in addition to other structural studies on whole enzyme and related proteins, the research team also performed experiments to elucidate the mechanisms of the enzyme reaction, validating detailed structures, enzyme engineering, and also N-terminal domain studies among others.
Through several biochemical studies based on crystal structure, the authors show that PHA synthase exists as a dimer and is divided into two distinct domains, the N-terminal domain (RePhaC1ND) and the C-terminal domain (RePhaC1CD). The RePhaC1CD catalyzes the polymerization reaction via a non-processive ping-pong mechanism using a Cys-His-Asp catalytic triad. The two catalytic sites of the RePhaC1CD dimer are positioned 33.4 Å apart, suggesting that the polymerization reaction occurs independently at each site. This study also presents the structure-based mechanisms for substrate specificities of various PHA synthases from different classes.
Professor Sang Yup Lee, who has worked on this topic for more than 20 years, said,
“The results and information presented in these two papers have long been awaited not only in the PHA community, but also metabolic engineering, bacteriology/microbiology, and in general biological sciences communities. The structural information on PHA synthase together with the recently deciphered reaction mechanisms will be valuable for understanding the detailed mechanisms of biosynthesizing this important energy/redox storage material, and also for the rational engineering of PHA synthases to produce designer bioplastics from various monomers more efficiently.”
Indeed, these two papers published in Biotechnology Journal finally reveal the 30-year mystery of machinery of biological polyester synthesis, and will serve as the essential compass in creating designer and more efficient bioplastic machineries.
References:
Jieun Kim, Yeo-Jin Kim, So Young Choi, Sang Yup Lee and Kyung-Jin Kim. “Crystal structure of Ralstonia eutropha polyhydroxyalkanoate synthase C-terminal domain and reaction mechanisms” Biotechnology Journal DOI: 10.1002/biot.201600648
http://onlinelibrary.wiley.com/doi/10.1002/biot.201600648/abstract
Yeo-Jin Kim, So Young Choi, Jieun Kim, Kyeong Sik Jin, Sang Yup Lee and Kyung-Jin Kim. “Structure and function of the N-terminal domain of Ralstonia eutropha polyhydroxyalkanoate synthase, and the proposed structure and mechanisms of the whole enzyme” Biotechnology Journal DOI: 10.1002/biot.201600649
http://onlinelibrary.wiley.com/doi/10.1002/biot.201600649/abstract
2016.12.02 View 12149 -
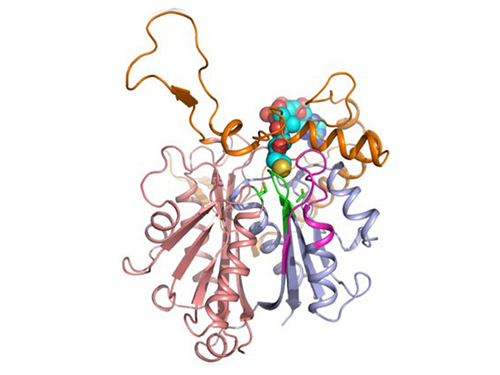 Discovery of Redox-Switch of KEenzyme Involved in N-Butanol Biosynthesis
Research teams at KAIST and Kyungpook National University (KNU) have succeeded in uncovering the redox-switch of thiolase, a key enzyme for n-butanol production in Clostridium acetobutylicum, one of the best known butanol-producing bacteria.
Biological n-butanol production was first reported by Louis Pasteur in 1861, and the bioprocess was industrialized usingClostridium acetobutylicum. The fermentation process by Clostridium strains has been known to be the most efficient one for n-butanol production. Due to growing world-wide issues such as energy security and climate change, the biological production of n-butanol has been receiving much renewed interest. This is because n-butanol possesses much better fuel characteristics compared to ethanol, such as higher energy content (29.2 MJ/L vs 19.6 MJ/L), less corrosiveness, less hygroscopy, and the ease with which it can be blended with gasoline and diesel.
In the paper published in Nature Communications, a broad-scope, online-only, and open access journal issued by the Nature Publishing Group (NPG), on September 22, 2015, Professor Kyung-Jin Kim at the School of Life Sciences, KNU, and Distinguished Professor Sang Yup Lee at the Department of Chemical and Biomolecular Engineering, KAIST, have proved that the redox-switch of thiolase plays a role in a regulation of metabolic flux in C. acetobutylicum by using in silico modeling and simulation tools.
The research team has redesigned thiolase with enhanced activity on the basis of the 3D structure of the wild-type enzyme. To reinforce a metabolic flux toward butanol production, the metabolic network of C. acetobutylicum strain was engineered with the redesigned enzyme. The combination of the discovery of 3D enzyme structure and systems metabolic engineering approaches resulted in increased n-butanol production in C. acetobutylicum, which allows the production of this important industrial chemical to be cost competitive.
Professors Kim and Lee said, "We have reported the 3D structure of C. acetobutylicum thiolase-a key enzyme involved in n-butanol biosynthesis, for the first time. Further study will be done to produce butanol more economically on the basis of the 3D structure of C. acetobutylicum thiolase."
This work was published online in Nature Communications on September 22, 2015.
Reference: Kim et al. "Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum," Nature Communications
This research was supported by the Technology Development Program to Solve Climate Changes from the Ministry of Education, Science and Technology (MEST), Korea, the National Research Foundation of Korea, and the Advanced Biomass Center through the Global Frontier Research Program of the MEST, Korea.
For further information, contact Dr. Sang Yup Lee, Distinguished Professor, KAIST, Daejeon, Korea (leesy@kaist.ac.kr, +82-42-350-3930); and Dr. Kyung-Jin Kim, Professor, KNU, Daegu, Korea (kkim@knu.ac.kr, +82-53-950-6088).
Figure 1: A redox-switch of thiolase involves in butanol biosynthesis in Clostridium acetobutylicum. Thiolase condenses two acetyl-CoA molecules for initiating four carbon flux towards butanol.
Figure 2: Thiolase catalyzes the condensation reaction of acetyl-CoA to acetoacetyl-CoA. Two catalytic cysteine residues at 88th and 378th are oxidized and formed an intermolecular disulfide bond in an oxidized status, which results in inactivation of the enzyme for n-butanol biosynthesis. The intermolecular disulfide bond is broken enabling the n-butanol biosynthesis, when the environment status is reduced.
2015.09.23 View 12342
Discovery of Redox-Switch of KEenzyme Involved in N-Butanol Biosynthesis
Research teams at KAIST and Kyungpook National University (KNU) have succeeded in uncovering the redox-switch of thiolase, a key enzyme for n-butanol production in Clostridium acetobutylicum, one of the best known butanol-producing bacteria.
Biological n-butanol production was first reported by Louis Pasteur in 1861, and the bioprocess was industrialized usingClostridium acetobutylicum. The fermentation process by Clostridium strains has been known to be the most efficient one for n-butanol production. Due to growing world-wide issues such as energy security and climate change, the biological production of n-butanol has been receiving much renewed interest. This is because n-butanol possesses much better fuel characteristics compared to ethanol, such as higher energy content (29.2 MJ/L vs 19.6 MJ/L), less corrosiveness, less hygroscopy, and the ease with which it can be blended with gasoline and diesel.
In the paper published in Nature Communications, a broad-scope, online-only, and open access journal issued by the Nature Publishing Group (NPG), on September 22, 2015, Professor Kyung-Jin Kim at the School of Life Sciences, KNU, and Distinguished Professor Sang Yup Lee at the Department of Chemical and Biomolecular Engineering, KAIST, have proved that the redox-switch of thiolase plays a role in a regulation of metabolic flux in C. acetobutylicum by using in silico modeling and simulation tools.
The research team has redesigned thiolase with enhanced activity on the basis of the 3D structure of the wild-type enzyme. To reinforce a metabolic flux toward butanol production, the metabolic network of C. acetobutylicum strain was engineered with the redesigned enzyme. The combination of the discovery of 3D enzyme structure and systems metabolic engineering approaches resulted in increased n-butanol production in C. acetobutylicum, which allows the production of this important industrial chemical to be cost competitive.
Professors Kim and Lee said, "We have reported the 3D structure of C. acetobutylicum thiolase-a key enzyme involved in n-butanol biosynthesis, for the first time. Further study will be done to produce butanol more economically on the basis of the 3D structure of C. acetobutylicum thiolase."
This work was published online in Nature Communications on September 22, 2015.
Reference: Kim et al. "Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum," Nature Communications
This research was supported by the Technology Development Program to Solve Climate Changes from the Ministry of Education, Science and Technology (MEST), Korea, the National Research Foundation of Korea, and the Advanced Biomass Center through the Global Frontier Research Program of the MEST, Korea.
For further information, contact Dr. Sang Yup Lee, Distinguished Professor, KAIST, Daejeon, Korea (leesy@kaist.ac.kr, +82-42-350-3930); and Dr. Kyung-Jin Kim, Professor, KNU, Daegu, Korea (kkim@knu.ac.kr, +82-53-950-6088).
Figure 1: A redox-switch of thiolase involves in butanol biosynthesis in Clostridium acetobutylicum. Thiolase condenses two acetyl-CoA molecules for initiating four carbon flux towards butanol.
Figure 2: Thiolase catalyzes the condensation reaction of acetyl-CoA to acetoacetyl-CoA. Two catalytic cysteine residues at 88th and 378th are oxidized and formed an intermolecular disulfide bond in an oxidized status, which results in inactivation of the enzyme for n-butanol biosynthesis. The intermolecular disulfide bond is broken enabling the n-butanol biosynthesis, when the environment status is reduced.
2015.09.23 View 12342