BEL
-
 KAIST and Mainz Researchers Unveil 3D Magnon Control, Charting a New Course for Neuromorphic and Quantum Technologies
< Professor Se Kwon Kim of the Department of Physics (left), Dr. Zarzuela of the University of Mainz, Germany (right) >
What if the magnon Hall effect, which processes information using magnons (spin waves) capable of current-free information transfer with magnets, could overcome its current limitation of being possible only on a 2D plane? If magnons could be utilized in 3D space, they would enable flexible design, including 3D circuits, and be applicable in various fields such as next-generation neuromorphic (brain-mimicking) computing structures, similar to human brain information processing. KAIST and an international joint research team have, for the first time in the world, predicted a 3D magnon Hall effect, demonstrating that magnons can move freely and complexly in 3D space, transcending the conventional concept of magnons.
KAIST (President Kwang Hyung Lee) announced on May 22nd that Professor Se Kwon Kim of the Department of Physics, in collaboration with Dr. Ricardo Zarzuela of the University of Mainz, Germany, has revealed that the interaction between magnons (spin waves) and solitons (spin vortices) within complex magnetic structures (topologically textured frustrated magnets) is not simple, but complex in a way that enables novel functionalities.
Magnons (spin waves), which can transmit information like electron movement, are garnering attention as a next-generation information processing technology that transmits information without using current, thus generating no heat. Until now, magnon research has focused on simple magnets where spins are neatly aligned in one direction, and the mathematics describing this was a relatively simple 'Abelian gauge theory.'
The research team demonstrated, for the first time in the world, that in complex spin structures like frustrated magnets, magnons interact and become entangled in complex ways from various directions. They applied an advanced mathematical framework, 'non-Abelian gauge theory,' to describe this movement, which is a groundbreaking achievement.
This research presents the possibility of future applications in low-power logic devices using magnons and topology-based quantum information processing technologies, indicating a potential paradigm shift in future information technology.
In conventional linear magnetic materials, the value representing the magnetic state (order parameter) is given as a vector. In magnonics research based on this, it has been interpreted that a U(1) Abelian gauge field is induced when magnons move in soliton structures like skyrmions. This means that the interaction between solitons and magnons has a structure similar to quantum electrodynamics (QED), which has successfully explained various experimental results such as the magnon Hall effect in 2D magnets.
< Figure. Schematic diagram of non-Abelian magnon quantum chromodynamics describing the dynamics of three types of magnons discovered for the first time in this study.>
However, through this research, the team theoretically revealed that in frustrated magnets, the order parameter must be expressed not as a simple vector but as a quaternion. As a result, the gauge field experienced by magnons resembles an SU(3) non-Abelian gauge field, rather than a simple U(1) Abelian gauge field.
This implies that within frustrated magnets, there are not one or two types of magnons seen in conventional magnets, but three distinct types of magnons, each interacting and intricately entangled with solitons. This structure is highly significant as it resembles quantum chromodynamics (QCD) that describes the strong interaction between quarks mediated by gluons rather than quantum electrodynamics (QED) that describes electromagnetic forces.
Professor Se Kwon Kim stated, "This research presents a powerful theoretical framework to explain the dynamics of magnons occurring within the complex order of frustrated magnets," adding, "By pioneering non-Abelian magnonics, it will be a conceptual turning point that can influence quantum magnetism research as a whole."
The research results, with Dr. Ricardo Zarzuela of the University of Mainz, Germany, as the first author, were published in the world-renowned physics journal Physical Review Letters on May 6th.※ Paper title: "Non-Abelian Gauge Theory for Magnons in Topologically Textured Frustrated Magnets," Phys. Rev. Lett. 134, 186701 (2025)DOI: https://doi.org/10.1103/PhysRevLett.134.186701
This research was supported by the Brain Pool Plus program of the National Research Foundation of Korea.
2025.05.22 View 4143
KAIST and Mainz Researchers Unveil 3D Magnon Control, Charting a New Course for Neuromorphic and Quantum Technologies
< Professor Se Kwon Kim of the Department of Physics (left), Dr. Zarzuela of the University of Mainz, Germany (right) >
What if the magnon Hall effect, which processes information using magnons (spin waves) capable of current-free information transfer with magnets, could overcome its current limitation of being possible only on a 2D plane? If magnons could be utilized in 3D space, they would enable flexible design, including 3D circuits, and be applicable in various fields such as next-generation neuromorphic (brain-mimicking) computing structures, similar to human brain information processing. KAIST and an international joint research team have, for the first time in the world, predicted a 3D magnon Hall effect, demonstrating that magnons can move freely and complexly in 3D space, transcending the conventional concept of magnons.
KAIST (President Kwang Hyung Lee) announced on May 22nd that Professor Se Kwon Kim of the Department of Physics, in collaboration with Dr. Ricardo Zarzuela of the University of Mainz, Germany, has revealed that the interaction between magnons (spin waves) and solitons (spin vortices) within complex magnetic structures (topologically textured frustrated magnets) is not simple, but complex in a way that enables novel functionalities.
Magnons (spin waves), which can transmit information like electron movement, are garnering attention as a next-generation information processing technology that transmits information without using current, thus generating no heat. Until now, magnon research has focused on simple magnets where spins are neatly aligned in one direction, and the mathematics describing this was a relatively simple 'Abelian gauge theory.'
The research team demonstrated, for the first time in the world, that in complex spin structures like frustrated magnets, magnons interact and become entangled in complex ways from various directions. They applied an advanced mathematical framework, 'non-Abelian gauge theory,' to describe this movement, which is a groundbreaking achievement.
This research presents the possibility of future applications in low-power logic devices using magnons and topology-based quantum information processing technologies, indicating a potential paradigm shift in future information technology.
In conventional linear magnetic materials, the value representing the magnetic state (order parameter) is given as a vector. In magnonics research based on this, it has been interpreted that a U(1) Abelian gauge field is induced when magnons move in soliton structures like skyrmions. This means that the interaction between solitons and magnons has a structure similar to quantum electrodynamics (QED), which has successfully explained various experimental results such as the magnon Hall effect in 2D magnets.
< Figure. Schematic diagram of non-Abelian magnon quantum chromodynamics describing the dynamics of three types of magnons discovered for the first time in this study.>
However, through this research, the team theoretically revealed that in frustrated magnets, the order parameter must be expressed not as a simple vector but as a quaternion. As a result, the gauge field experienced by magnons resembles an SU(3) non-Abelian gauge field, rather than a simple U(1) Abelian gauge field.
This implies that within frustrated magnets, there are not one or two types of magnons seen in conventional magnets, but three distinct types of magnons, each interacting and intricately entangled with solitons. This structure is highly significant as it resembles quantum chromodynamics (QCD) that describes the strong interaction between quarks mediated by gluons rather than quantum electrodynamics (QED) that describes electromagnetic forces.
Professor Se Kwon Kim stated, "This research presents a powerful theoretical framework to explain the dynamics of magnons occurring within the complex order of frustrated magnets," adding, "By pioneering non-Abelian magnonics, it will be a conceptual turning point that can influence quantum magnetism research as a whole."
The research results, with Dr. Ricardo Zarzuela of the University of Mainz, Germany, as the first author, were published in the world-renowned physics journal Physical Review Letters on May 6th.※ Paper title: "Non-Abelian Gauge Theory for Magnons in Topologically Textured Frustrated Magnets," Phys. Rev. Lett. 134, 186701 (2025)DOI: https://doi.org/10.1103/PhysRevLett.134.186701
This research was supported by the Brain Pool Plus program of the National Research Foundation of Korea.
2025.05.22 View 4143 -
 KAIST sends out Music and Bio-Signs of Professor Kwon Ji-yong, a.k.a. G-Dragon, into Space to Pulsate through Universe and Resonate among Stars
KAIST (President Kwang-Hyung Lee) announced on the 10th of April that it successfully promoted the world’s first ‘Space Sound Source Transmission Project’ based on media art at the KAIST Space Research Institute on April 9th through collaboration between Professor Jinjoon Lee of the Graduate School of Culture Technology, a world-renowned media artist, and the global K-Pop artist, G-Dragon.
This project was proposed as part of the ‘AI Entertech Research Center’ being promoted by KAIST and Galaxy Corporation. It is a project to transmit the message and sound of G-Dragon (real name, Kwon Ji-yong), a singer/song writer affiliated with Galaxy Corporation and a visiting professor in the Department of Mechanical Engineering at KAIST, to space for the first time in the world.
This is a convergence project that combines science, technology, art, and popular music, and is a new form of ‘space culture content’ experiment that connects KAIST’s cutting-edge space technology, Professor Jinjoon Lee’s media art work, and G-Dragon’s voice and sound source containing his latest digital single, "HOME SWEET HOME".
< Photo 1. Professor Jinjoon Lee's Open Your Eyes Project "Iris"'s imagery projected on the 13m space antenna at the Space Research Institute >
This collaboration was planned with the theme of ‘emotional signals that expand the inner universe of humans to the outer universe.’ The image of G-Dragon’s iris was augmented through AI as a window into soul symbolizing his uniqueness and identity, and the new song “Home Sweet Home” was combined as an audio message containing the vibration of that emotion.
This was actually transmitted into space using a next-generation small satellite developed by KAIST Space Research Institute, completing a symbolic performance in which an individual’s inner universe is transmitted to outer space.
Professor Jinjoon Lee’s cinematic media art work “Iris” was unveiled at the site. This work was screened in the world’s first projection mapping method* on KAIST Space Research Institute’s 13m space antenna. This video was created using generative artificial intelligence (AI) technology based on the image of G-Dragon's iris, and combined with sound using the data of the sounds of Emile Bell rings – the bell that holds a thousand years of history, it presented an emotional art experience that transcends time and space.
*Projection Mapping: A technology that projects light and images onto actual structures to create visual changes, and is a method of expression that artistically reinterprets space.
This work is one of the major research achievements of KAIST TX Lab and Professor Lee based on new media technology based on biometric data such as iris, heartbeat, and brain waves.
Professor Jinjoon Lee said, "The iris is a symbol that reflects inner emotions and identity, so much so that it is called the 'mirror of the soul,' and this work sought to express 'the infinite universe seen from the inside of humanity' through G-Dragon's gaze."
< Photo 2. (From left) Professor Jinjoon Lee of the Graduate School of Culture Technology and G-Dragon (Visiting Professor Kwon Ji-yong of the Department of Mechanical Engineering) >
He continued, "The universe is a realm of technology as well as a stage for imagination and emotion, and I look forward to an encounter with the unknown through a new attempt to speak of art in the language of science including AI and imagine science in the form of art." “G-Dragon’s voice and music have now begun their journey to space,” said Yong-ho Choi, Galaxy Corporation’s Chief Happiness Officer (CHO). “This project is an act of leaving music as a legacy for humanity, while also having an important meaning of attempting to communicate with space.” He added, “This is a pioneering step to introduce human culture to space, and it will remain as a monumental performance that opens a new chapter in the history of music comparable to the Beatles.”
Galaxy Corporation is leading the future entertainment technology industry through its collaboration with KAIST, and was recently selected as the only entertainment technology company in a private meeting with Microsoft CEO Nadella. In particular, it is promoting the globalization of AI entertainment technology, receiving praise as a “pioneer of imagination” for new forms of AI entertainment content, including the AI contents for the deceased.
< Photo 3. Photo of G-Dragon's Home Sweet Home being sent into the space via Professor Jinjoon Lee's Space Sound Source Transmission Project >
Through this project, KAIST Space Research Institute presented new possibilities for utilizing satellite technology, and showed a model for science to connect with society in a more popular way.
KAIST President Kwang-Hyung Lee said, “KAIST is a place that always supports new imaginations and challenges,” and added, “We will continue to strive to continue creative research that no one has ever thought of, like this project that combines science, technology, and art.”
In the meantime, Galaxy Corporation, the agency of G-Dragon’s Professor Kwon Ji-yong, is an AI entertainment company that presents a new paradigm based on IP, media, tech, and entertainment convergence technology.
2025.04.10 View 6192
KAIST sends out Music and Bio-Signs of Professor Kwon Ji-yong, a.k.a. G-Dragon, into Space to Pulsate through Universe and Resonate among Stars
KAIST (President Kwang-Hyung Lee) announced on the 10th of April that it successfully promoted the world’s first ‘Space Sound Source Transmission Project’ based on media art at the KAIST Space Research Institute on April 9th through collaboration between Professor Jinjoon Lee of the Graduate School of Culture Technology, a world-renowned media artist, and the global K-Pop artist, G-Dragon.
This project was proposed as part of the ‘AI Entertech Research Center’ being promoted by KAIST and Galaxy Corporation. It is a project to transmit the message and sound of G-Dragon (real name, Kwon Ji-yong), a singer/song writer affiliated with Galaxy Corporation and a visiting professor in the Department of Mechanical Engineering at KAIST, to space for the first time in the world.
This is a convergence project that combines science, technology, art, and popular music, and is a new form of ‘space culture content’ experiment that connects KAIST’s cutting-edge space technology, Professor Jinjoon Lee’s media art work, and G-Dragon’s voice and sound source containing his latest digital single, "HOME SWEET HOME".
< Photo 1. Professor Jinjoon Lee's Open Your Eyes Project "Iris"'s imagery projected on the 13m space antenna at the Space Research Institute >
This collaboration was planned with the theme of ‘emotional signals that expand the inner universe of humans to the outer universe.’ The image of G-Dragon’s iris was augmented through AI as a window into soul symbolizing his uniqueness and identity, and the new song “Home Sweet Home” was combined as an audio message containing the vibration of that emotion.
This was actually transmitted into space using a next-generation small satellite developed by KAIST Space Research Institute, completing a symbolic performance in which an individual’s inner universe is transmitted to outer space.
Professor Jinjoon Lee’s cinematic media art work “Iris” was unveiled at the site. This work was screened in the world’s first projection mapping method* on KAIST Space Research Institute’s 13m space antenna. This video was created using generative artificial intelligence (AI) technology based on the image of G-Dragon's iris, and combined with sound using the data of the sounds of Emile Bell rings – the bell that holds a thousand years of history, it presented an emotional art experience that transcends time and space.
*Projection Mapping: A technology that projects light and images onto actual structures to create visual changes, and is a method of expression that artistically reinterprets space.
This work is one of the major research achievements of KAIST TX Lab and Professor Lee based on new media technology based on biometric data such as iris, heartbeat, and brain waves.
Professor Jinjoon Lee said, "The iris is a symbol that reflects inner emotions and identity, so much so that it is called the 'mirror of the soul,' and this work sought to express 'the infinite universe seen from the inside of humanity' through G-Dragon's gaze."
< Photo 2. (From left) Professor Jinjoon Lee of the Graduate School of Culture Technology and G-Dragon (Visiting Professor Kwon Ji-yong of the Department of Mechanical Engineering) >
He continued, "The universe is a realm of technology as well as a stage for imagination and emotion, and I look forward to an encounter with the unknown through a new attempt to speak of art in the language of science including AI and imagine science in the form of art." “G-Dragon’s voice and music have now begun their journey to space,” said Yong-ho Choi, Galaxy Corporation’s Chief Happiness Officer (CHO). “This project is an act of leaving music as a legacy for humanity, while also having an important meaning of attempting to communicate with space.” He added, “This is a pioneering step to introduce human culture to space, and it will remain as a monumental performance that opens a new chapter in the history of music comparable to the Beatles.”
Galaxy Corporation is leading the future entertainment technology industry through its collaboration with KAIST, and was recently selected as the only entertainment technology company in a private meeting with Microsoft CEO Nadella. In particular, it is promoting the globalization of AI entertainment technology, receiving praise as a “pioneer of imagination” for new forms of AI entertainment content, including the AI contents for the deceased.
< Photo 3. Photo of G-Dragon's Home Sweet Home being sent into the space via Professor Jinjoon Lee's Space Sound Source Transmission Project >
Through this project, KAIST Space Research Institute presented new possibilities for utilizing satellite technology, and showed a model for science to connect with society in a more popular way.
KAIST President Kwang-Hyung Lee said, “KAIST is a place that always supports new imaginations and challenges,” and added, “We will continue to strive to continue creative research that no one has ever thought of, like this project that combines science, technology, and art.”
In the meantime, Galaxy Corporation, the agency of G-Dragon’s Professor Kwon Ji-yong, is an AI entertainment company that presents a new paradigm based on IP, media, tech, and entertainment convergence technology.
2025.04.10 View 6192 -
 KAIST Develops CamBio - a New Biotemplating Method
- Professor Jae-Byum Chang and Professor Yeon Sik Jung’s joint research team of the Department of Materials Science and Engineering developed a highly tunable bio-templating method “CamBio” that makes use of intracellular protein structures
- Substrate performance improvement of up to 230% demonstrated via surface-enhanced Raman spectroscopy (SERS)
- Expected to have price competitiveness over bio-templating method as it expands the range of biological samples
- Expected to expand the range of application of nanostructure synthesis technology by utilizing various biological structures
< Photo 1. (From left) Professor Yeon Sik Jung, Ph.D. candidate Dae-Hyeon Song, Professor Jae-Byum Chang, and (from top right) Dr. Chang Woo Song and Dr. Seunghee H. Cho of the Department of Materials Science and Engineering >
Biological structures have complex characteristics that are difficult to replicate artificially, but biotemplating methods* that directly utilize these biological structures have been used in various fields of application. The KAIST research team succeeded in utilizing previously unusable biological structures and expanding the areas in which biotemplate methods can be applied.
*Biotemplating: A method of using biotemplates as a mold to create functional structural materials, utilizing the functions of these biological structures, from viruses to the tissues and organs that make up our bodies
KAIST (President Kwang Hyung Lee) announced on the 10th that a joint research team of Professors Jae-Byum Chang and Professor Yeon Sik Jung of the Department of Materials Science and Engineering developed a biotemplating method that utilizes specific intracellular proteins in biological samples and has high tunability.
Existing biotemplate methods mainly utilize only the external surface of biological samples or have limitations in utilizing the structure-function correlation of various biological structures due to limited dimensions and sample sizes, making it difficult to create functional nanostructures.
To solve this problem, the research team studied a way to utilize various biological structures within the cells while retaining high tunability.
< Figure 1. CamBio utilizing microtubules, a intracellular protein structure. The silver nanoparticle chains synthesized along the microtubules that span the entire cell interior can be observed through an electron microscope, and it is shown that this can be used as a successful SERS substrate. >
As a result of the research, the team developed the “Conversion to advanced materials via labeled Biostructure”, shortened as “CamBio”, which enables the selective synthesis of nanostructures with various characteristics and sizes from specific protein structures composed of diverse proteins within biological specimens.
The CamBio method secures high tunability of functional nanostructures that can be manufactured from biological samples by merging various manufacturing and biological technologies.
Through the technology of repeatedly attaching antibodies, arranging cells in a certain shape, and thinly slicing tissue, the functional nanostructures made with CamBio showed improved performance on the surface-enhanced Raman spectroscopy (SERS)* substrate used for material detection.
*Surface-enhanced Raman spectroscopy (SERS): A technology that can detect very small amounts of substances using light, based on the principle that specific substances react to light and amplifies signals on surfaces of metals such as gold or silver.
The research team found that the nanoparticle chains made using the intracellular protein structures through the process of repeated labeling with antibodies allowed easier control, and improved SERS performance by up to 230%.
In addition, the research team expanded from utilizing the structures inside cells to obtaining samples of muscle tissues inside meat using a cryostat and successfully producing a substrate with periodic bands made of metal particles by performing the CamBio process. This method of producing a substrate not only allows large-scale production using biological samples, but also shows that it is a cost-effective method.
< Figure 2. A method for securing tunability using CamBio at the cell level. Examples of controlling characteristics by integrating iterative labeling and cell pattering techniques with CamBio are shown. >
The CamBio developed by the research team is expected to be used as a way to solve problems faced by various research fields as it is to expand the range of bio-samples that can be produced for various usage.
The first author, Dae-Hyeon Song, a Ph.D. candidate of KAIST Department of Materials Science and Engineering said, “Through CamBio, we have comprehensively accumulated biotemplating methods that can utilize more diverse protein structures,” and “If combined with the state-of-the-art biological technologies such as gene editing and 3D bioprinting and new material synthesis technologies, biostructures can be utilized in various fields of application.”
< Figure 3. A method for securing tunability using CamBio at the tissue level. In order to utilize proteins inside muscle tissue, the frozen tissue sectioning technology is combined, and through this, a substrate with a periodic nanoparticle band pattern is successfully produced, and it is shown that large-area acquisition of samples and price competitiveness can be achieved. >
This study, in which the Ph.D. candidate Dae-Hyeon Song along with Dr. Chang Woo Song, and Dr. Seunghee H. Cho of the same department participated as the first authors, was published online in the international academic journal, Advanced Science, on November 13th, 2024.
(Paper title: Highly Tunable, Nanomaterial-Functionalized Structural Templating of Intracellular Protein Structures Within Biological Species) https://doi.org/10.1002/advs.202406492
This study was conducted with a combination of support from various programs including the National Convergence Research of Scientific Challenges (National Research Foundation of Korea (NRF) 2024), Engineering Reseach Center (ERC) (Wearable Platform Materials Technology Center, NRF 2023), ERC (Global Bio-integrated Materials Center, NRF 2024), and the National Advanced Program for Biological Research Resources (Bioimaging Data Curation Center, NRF 2024) funded by Ministry of Science and ICT.
2025.01.10 View 6679
KAIST Develops CamBio - a New Biotemplating Method
- Professor Jae-Byum Chang and Professor Yeon Sik Jung’s joint research team of the Department of Materials Science and Engineering developed a highly tunable bio-templating method “CamBio” that makes use of intracellular protein structures
- Substrate performance improvement of up to 230% demonstrated via surface-enhanced Raman spectroscopy (SERS)
- Expected to have price competitiveness over bio-templating method as it expands the range of biological samples
- Expected to expand the range of application of nanostructure synthesis technology by utilizing various biological structures
< Photo 1. (From left) Professor Yeon Sik Jung, Ph.D. candidate Dae-Hyeon Song, Professor Jae-Byum Chang, and (from top right) Dr. Chang Woo Song and Dr. Seunghee H. Cho of the Department of Materials Science and Engineering >
Biological structures have complex characteristics that are difficult to replicate artificially, but biotemplating methods* that directly utilize these biological structures have been used in various fields of application. The KAIST research team succeeded in utilizing previously unusable biological structures and expanding the areas in which biotemplate methods can be applied.
*Biotemplating: A method of using biotemplates as a mold to create functional structural materials, utilizing the functions of these biological structures, from viruses to the tissues and organs that make up our bodies
KAIST (President Kwang Hyung Lee) announced on the 10th that a joint research team of Professors Jae-Byum Chang and Professor Yeon Sik Jung of the Department of Materials Science and Engineering developed a biotemplating method that utilizes specific intracellular proteins in biological samples and has high tunability.
Existing biotemplate methods mainly utilize only the external surface of biological samples or have limitations in utilizing the structure-function correlation of various biological structures due to limited dimensions and sample sizes, making it difficult to create functional nanostructures.
To solve this problem, the research team studied a way to utilize various biological structures within the cells while retaining high tunability.
< Figure 1. CamBio utilizing microtubules, a intracellular protein structure. The silver nanoparticle chains synthesized along the microtubules that span the entire cell interior can be observed through an electron microscope, and it is shown that this can be used as a successful SERS substrate. >
As a result of the research, the team developed the “Conversion to advanced materials via labeled Biostructure”, shortened as “CamBio”, which enables the selective synthesis of nanostructures with various characteristics and sizes from specific protein structures composed of diverse proteins within biological specimens.
The CamBio method secures high tunability of functional nanostructures that can be manufactured from biological samples by merging various manufacturing and biological technologies.
Through the technology of repeatedly attaching antibodies, arranging cells in a certain shape, and thinly slicing tissue, the functional nanostructures made with CamBio showed improved performance on the surface-enhanced Raman spectroscopy (SERS)* substrate used for material detection.
*Surface-enhanced Raman spectroscopy (SERS): A technology that can detect very small amounts of substances using light, based on the principle that specific substances react to light and amplifies signals on surfaces of metals such as gold or silver.
The research team found that the nanoparticle chains made using the intracellular protein structures through the process of repeated labeling with antibodies allowed easier control, and improved SERS performance by up to 230%.
In addition, the research team expanded from utilizing the structures inside cells to obtaining samples of muscle tissues inside meat using a cryostat and successfully producing a substrate with periodic bands made of metal particles by performing the CamBio process. This method of producing a substrate not only allows large-scale production using biological samples, but also shows that it is a cost-effective method.
< Figure 2. A method for securing tunability using CamBio at the cell level. Examples of controlling characteristics by integrating iterative labeling and cell pattering techniques with CamBio are shown. >
The CamBio developed by the research team is expected to be used as a way to solve problems faced by various research fields as it is to expand the range of bio-samples that can be produced for various usage.
The first author, Dae-Hyeon Song, a Ph.D. candidate of KAIST Department of Materials Science and Engineering said, “Through CamBio, we have comprehensively accumulated biotemplating methods that can utilize more diverse protein structures,” and “If combined with the state-of-the-art biological technologies such as gene editing and 3D bioprinting and new material synthesis technologies, biostructures can be utilized in various fields of application.”
< Figure 3. A method for securing tunability using CamBio at the tissue level. In order to utilize proteins inside muscle tissue, the frozen tissue sectioning technology is combined, and through this, a substrate with a periodic nanoparticle band pattern is successfully produced, and it is shown that large-area acquisition of samples and price competitiveness can be achieved. >
This study, in which the Ph.D. candidate Dae-Hyeon Song along with Dr. Chang Woo Song, and Dr. Seunghee H. Cho of the same department participated as the first authors, was published online in the international academic journal, Advanced Science, on November 13th, 2024.
(Paper title: Highly Tunable, Nanomaterial-Functionalized Structural Templating of Intracellular Protein Structures Within Biological Species) https://doi.org/10.1002/advs.202406492
This study was conducted with a combination of support from various programs including the National Convergence Research of Scientific Challenges (National Research Foundation of Korea (NRF) 2024), Engineering Reseach Center (ERC) (Wearable Platform Materials Technology Center, NRF 2023), ERC (Global Bio-integrated Materials Center, NRF 2024), and the National Advanced Program for Biological Research Resources (Bioimaging Data Curation Center, NRF 2024) funded by Ministry of Science and ICT.
2025.01.10 View 6679 -
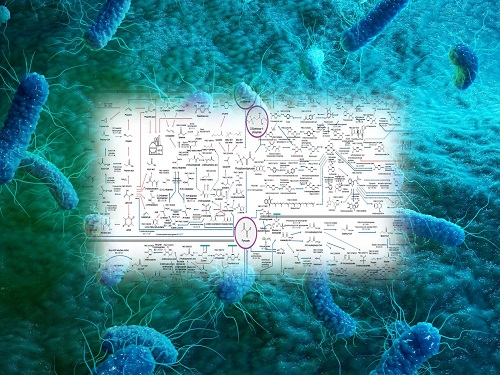 Interactive Map of Metabolical Synthesis of Chemicals
An interactive map that compiled the chemicals produced by biological, chemical and combined reactions has been distributed on the web
- A team led by Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering, organized and distributed an all-inclusive listing of chemical substances that can be synthesized using microorganisms
- It is expected to be used by researchers around the world as it enables easy assessment of the synthetic pathway through the web.
A research team comprised of Woo Dae Jang, Gi Bae Kim, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST reported an interactive metabolic map of bio-based chemicals. Their research paper “An interactive metabolic map of bio-based chemicals” was published online in Trends in Biotechnology on August 10, 2022.
As a response to rapid climate change and environmental pollution, research on the production of petrochemical products using microorganisms is receiving attention as a sustainable alternative to existing methods of productions. In order to synthesize various chemical substances, materials, and fuel using microorganisms, it is necessary to first construct the biosynthetic pathway toward desired product by exploration and discovery and introduce them into microorganisms. In addition, in order to efficiently synthesize various chemical substances, it is sometimes necessary to employ chemical methods along with bioengineering methods using microorganisms at the same time. For the production of non-native chemicals, novel pathways are designed by recruiting enzymes from heterologous sources or employing enzymes designed though rational engineering, directed evolution, or ab initio design.
The research team had completed a map of chemicals which compiled all available pathways of biological and/or chemical reactions that lead to the production of various bio-based chemicals back in 2019 and published the map in Nature Catalysis. The map was distributed in the form of a poster to industries and academia so that the synthesis paths of bio-based chemicals could be checked at a glance.
The research team has expanded the bio-based chemicals map this time in the form of an interactive map on the web so that anyone with internet access can quickly explore efficient paths to synthesize desired products. The web-based map provides interactive visual tools to allow interactive visualization, exploration, and analysis of complex networks of biological and/or chemical reactions toward the desired products. In addition, the reported paper also discusses the production of natural compounds that are used for diverse purposes such as food and medicine, which will help designing novel pathways through similar approaches or by exploiting the promiscuity of enzymes described in the map. The published bio-based chemicals map is also available at http://systemsbiotech.co.kr.
The co-first authors, Dr. Woo Dae Jang and Ph.D. student Gi Bae Kim, said, “We conducted this study to address the demand for updating the previously distributed chemicals map and enhancing its versatility.” “The map is expected to be utilized in a variety of research and in efforts to set strategies and prospects for chemical production incorporating bio and chemical methods that are detailed in the map.”
Distinguished Professor Sang Yup Lee said, “The interactive bio-based chemicals map is expected to help design and optimization of the metabolic pathways for the biosynthesis of target chemicals together with the strategies of chemical conversions, serving as a blueprint for developing further ideas on the production of desired chemicals through biological and/or chemical reactions.”
The interactive metabolic map of bio-based chemicals.
2022.08.11 View 18854
Interactive Map of Metabolical Synthesis of Chemicals
An interactive map that compiled the chemicals produced by biological, chemical and combined reactions has been distributed on the web
- A team led by Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering, organized and distributed an all-inclusive listing of chemical substances that can be synthesized using microorganisms
- It is expected to be used by researchers around the world as it enables easy assessment of the synthetic pathway through the web.
A research team comprised of Woo Dae Jang, Gi Bae Kim, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST reported an interactive metabolic map of bio-based chemicals. Their research paper “An interactive metabolic map of bio-based chemicals” was published online in Trends in Biotechnology on August 10, 2022.
As a response to rapid climate change and environmental pollution, research on the production of petrochemical products using microorganisms is receiving attention as a sustainable alternative to existing methods of productions. In order to synthesize various chemical substances, materials, and fuel using microorganisms, it is necessary to first construct the biosynthetic pathway toward desired product by exploration and discovery and introduce them into microorganisms. In addition, in order to efficiently synthesize various chemical substances, it is sometimes necessary to employ chemical methods along with bioengineering methods using microorganisms at the same time. For the production of non-native chemicals, novel pathways are designed by recruiting enzymes from heterologous sources or employing enzymes designed though rational engineering, directed evolution, or ab initio design.
The research team had completed a map of chemicals which compiled all available pathways of biological and/or chemical reactions that lead to the production of various bio-based chemicals back in 2019 and published the map in Nature Catalysis. The map was distributed in the form of a poster to industries and academia so that the synthesis paths of bio-based chemicals could be checked at a glance.
The research team has expanded the bio-based chemicals map this time in the form of an interactive map on the web so that anyone with internet access can quickly explore efficient paths to synthesize desired products. The web-based map provides interactive visual tools to allow interactive visualization, exploration, and analysis of complex networks of biological and/or chemical reactions toward the desired products. In addition, the reported paper also discusses the production of natural compounds that are used for diverse purposes such as food and medicine, which will help designing novel pathways through similar approaches or by exploiting the promiscuity of enzymes described in the map. The published bio-based chemicals map is also available at http://systemsbiotech.co.kr.
The co-first authors, Dr. Woo Dae Jang and Ph.D. student Gi Bae Kim, said, “We conducted this study to address the demand for updating the previously distributed chemicals map and enhancing its versatility.” “The map is expected to be utilized in a variety of research and in efforts to set strategies and prospects for chemical production incorporating bio and chemical methods that are detailed in the map.”
Distinguished Professor Sang Yup Lee said, “The interactive bio-based chemicals map is expected to help design and optimization of the metabolic pathways for the biosynthesis of target chemicals together with the strategies of chemical conversions, serving as a blueprint for developing further ideas on the production of desired chemicals through biological and/or chemical reactions.”
The interactive metabolic map of bio-based chemicals.
2022.08.11 View 18854 -
 Metabolically Engineered Bacterium Produces Lutein
A research group at KAIST has engineered a bacterial strain capable of producing lutein. The research team applied systems metabolic engineering strategies, including substrate channeling and electron channeling, to enhance the production of lutein in an engineered Escherichia coli strain. The strategies will be also useful for the efficient production of other industrially important natural products used in the food, pharmaceutical, and cosmetic industries.
Figure: Systems metabolic engineering was employed to construct and optimize the metabolic pathways for lutein production, and substrate channeling and electron channeling strategies were additionally employed to increase the production of the lutein with high productivity.
Lutein is classified as a xanthophyll chemical that is abundant in egg yolk, fruits, and vegetables. It protects the eye from oxidative damage from radiation and reduces the risk of eye diseases including macular degeneration and cataracts. Commercialized products featuring lutein are derived from the extracts of the marigold flower, which is known to harbor abundant amounts of lutein. However, the drawback of lutein production from nature is that it takes a long time to grow and harvest marigold flowers. Furthermore, it requires additional physical and chemical-based extractions with a low yield, which makes it economically unfeasible in terms of productivity. The high cost and low yield of these bioprocesses has made it difficult to readily meet the demand for lutein.
These challenges inspired the metabolic engineers at KAIST, including researchers Dr. Seon Young Park, Ph.D. Candidate Hyunmin Eun, and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering. The team’s study entitled “Metabolic engineering of Escherichia coli with electron channeling for the production of natural products” was published in Nature Catalysis on August 5, 2022.
This research details the ability to produce lutein from E. coli with a high yield using a cheap carbon source, glycerol, via systems metabolic engineering. The research group focused on solving the bottlenecks of the biosynthetic pathway for lutein production constructed within an individual cell. First, using systems metabolic engineering, which is an integrated technology to engineer the metabolism of a microorganism, lutein was produced when the lutein biosynthesis pathway was introduced, albeit in very small amounts.
To improve the productivity of lutein production, the bottleneck enzymes within the metabolic pathway were first identified. It turned out that metabolic reactions that involve a promiscuous enzyme, an enzyme that is involved in two or more metabolic reactions, and electron-requiring cytochrome P450 enzymes are the main bottleneck steps of the pathway inhibiting lutein biosynthesis.
To overcome these challenges, substrate channeling, a strategy to artificially recruit enzymes in physical proximity within the cell in order to increase the local concentrations of substrates that can be converted into products, was employed to channel more metabolic flux towards the target chemical while reducing the formation of unwanted byproducts.
Furthermore, electron channeling, a strategy similar to substrate channeling but differing in terms of increasing the local concentrations of electrons required for oxidoreduction reactions mediated by P450 and its reductase partners, was applied to further streamline the metabolic flux towards lutein biosynthesis, which led to the highest titer of lutein production achieved in a bacterial host ever reported. The same electron channeling strategy was successfully applied for the production of other natural products including nootkatone and apigenin in E. coli, showcasing the general applicability of the strategy in the research field.
“It is expected that this microbial cell factory-based production of lutein will be able to replace the current plant extraction-based process,” said Dr. Seon Young Park, the first author of the paper. She explained that another important point of the research is that integrated metabolic engineering strategies developed from this study can be generally applicable for the efficient production of other natural products useful as pharmaceuticals or nutraceuticals.
“As maintaining good health in an aging society is becoming increasingly important, we expect that the technology and strategies developed here will play pivotal roles in producing other valuable natural products of medical or nutritional importance,” explained Distinguished Professor Sang Yup Lee.
This work was supported by the Cooperative Research Program for Agriculture Science & Technology Development funded by the Rural Development Administration of Korea, with further support from the Development of Next-generation Biorefinery Platform Technologies for Leading Bio-based Chemicals Industry Project and by the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project of the National Research Foundation funded by the Ministry of Science and ICT of Korea.
2022.08.05 View 12510
Metabolically Engineered Bacterium Produces Lutein
A research group at KAIST has engineered a bacterial strain capable of producing lutein. The research team applied systems metabolic engineering strategies, including substrate channeling and electron channeling, to enhance the production of lutein in an engineered Escherichia coli strain. The strategies will be also useful for the efficient production of other industrially important natural products used in the food, pharmaceutical, and cosmetic industries.
Figure: Systems metabolic engineering was employed to construct and optimize the metabolic pathways for lutein production, and substrate channeling and electron channeling strategies were additionally employed to increase the production of the lutein with high productivity.
Lutein is classified as a xanthophyll chemical that is abundant in egg yolk, fruits, and vegetables. It protects the eye from oxidative damage from radiation and reduces the risk of eye diseases including macular degeneration and cataracts. Commercialized products featuring lutein are derived from the extracts of the marigold flower, which is known to harbor abundant amounts of lutein. However, the drawback of lutein production from nature is that it takes a long time to grow and harvest marigold flowers. Furthermore, it requires additional physical and chemical-based extractions with a low yield, which makes it economically unfeasible in terms of productivity. The high cost and low yield of these bioprocesses has made it difficult to readily meet the demand for lutein.
These challenges inspired the metabolic engineers at KAIST, including researchers Dr. Seon Young Park, Ph.D. Candidate Hyunmin Eun, and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering. The team’s study entitled “Metabolic engineering of Escherichia coli with electron channeling for the production of natural products” was published in Nature Catalysis on August 5, 2022.
This research details the ability to produce lutein from E. coli with a high yield using a cheap carbon source, glycerol, via systems metabolic engineering. The research group focused on solving the bottlenecks of the biosynthetic pathway for lutein production constructed within an individual cell. First, using systems metabolic engineering, which is an integrated technology to engineer the metabolism of a microorganism, lutein was produced when the lutein biosynthesis pathway was introduced, albeit in very small amounts.
To improve the productivity of lutein production, the bottleneck enzymes within the metabolic pathway were first identified. It turned out that metabolic reactions that involve a promiscuous enzyme, an enzyme that is involved in two or more metabolic reactions, and electron-requiring cytochrome P450 enzymes are the main bottleneck steps of the pathway inhibiting lutein biosynthesis.
To overcome these challenges, substrate channeling, a strategy to artificially recruit enzymes in physical proximity within the cell in order to increase the local concentrations of substrates that can be converted into products, was employed to channel more metabolic flux towards the target chemical while reducing the formation of unwanted byproducts.
Furthermore, electron channeling, a strategy similar to substrate channeling but differing in terms of increasing the local concentrations of electrons required for oxidoreduction reactions mediated by P450 and its reductase partners, was applied to further streamline the metabolic flux towards lutein biosynthesis, which led to the highest titer of lutein production achieved in a bacterial host ever reported. The same electron channeling strategy was successfully applied for the production of other natural products including nootkatone and apigenin in E. coli, showcasing the general applicability of the strategy in the research field.
“It is expected that this microbial cell factory-based production of lutein will be able to replace the current plant extraction-based process,” said Dr. Seon Young Park, the first author of the paper. She explained that another important point of the research is that integrated metabolic engineering strategies developed from this study can be generally applicable for the efficient production of other natural products useful as pharmaceuticals or nutraceuticals.
“As maintaining good health in an aging society is becoming increasingly important, we expect that the technology and strategies developed here will play pivotal roles in producing other valuable natural products of medical or nutritional importance,” explained Distinguished Professor Sang Yup Lee.
This work was supported by the Cooperative Research Program for Agriculture Science & Technology Development funded by the Rural Development Administration of Korea, with further support from the Development of Next-generation Biorefinery Platform Technologies for Leading Bio-based Chemicals Industry Project and by the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project of the National Research Foundation funded by the Ministry of Science and ICT of Korea.
2022.08.05 View 12510 -
 Tomographic Measurement of Dielectric Tensors
Dielectric tensor tomography allows the direct measurement of the 3D dielectric tensors of optically anisotropic structures
A research team reported the direct measurement of dielectric tensors of anisotropic structures including the spatial variations of principal refractive indices and directors. The group also demonstrated quantitative tomographic measurements of various nematic liquid-crystal structures and their fast 3D nonequilibrium dynamics using a 3D label-free tomographic method. The method was described in Nature Materials.
Light-matter interactions are described by the dielectric tensor. Despite their importance in basic science and applications, it has not been possible to measure 3D dielectric tensors directly. The main challenge was due to the vectorial nature of light scattering from a 3D anisotropic structure. Previous approaches only addressed 3D anisotropic information indirectly and were limited to two-dimensional, qualitative, strict sample conditions or assumptions.
The research team developed a method enabling the tomographic reconstruction of 3D dielectric tensors without any preparation or assumptions. A sample is illuminated with a laser beam with various angles and circularly polarization states. Then, the light fields scattered from a sample are holographically measured and converted into vectorial diffraction components. Finally, by inversely solving a vectorial wave equation, the 3D dielectric tensor is reconstructed.
Professor YongKeun Park said, “There were a greater number of unknowns in direct measuring than with the conventional approach. We applied our approach to measure additional holographic images by slightly tilting the incident angle.”
He said that the slightly tilted illumination provides an additional orthogonal polarization, which makes the underdetermined problem become the determined problem. “Although scattered fields are dependent on the illumination angle, the Fourier differentiation theorem enables the extraction of the same dielectric tensor for the slightly tilted illumination,” Professor Park added.
His team’s method was validated by reconstructing well-known liquid crystal (LC) structures, including the twisted nematic, hybrid aligned nematic, radial, and bipolar configurations. Furthermore, the research team demonstrated the experimental measurements of the non-equilibrium dynamics of annihilating, nucleating, and merging LC droplets, and the LC polymer network with repeating 3D topological defects.
“This is the first experimental measurement of non-equilibrium dynamics and 3D topological defects in LC structures in a label-free manner. Our method enables the exploration of inaccessible nematic structures and interactions in non-equilibrium dynamics,” first author Dr. Seungwoo Shin explained.
-PublicationSeungwoo Shin, Jonghee Eun, Sang Seok Lee, Changjae Lee, Herve Hugonnet, Dong Ki Yoon, Shin-Hyun Kim, Jongwoo Jeong, YongKeun Park, “Tomographic Measurement ofDielectric Tensors at Optical Frequency,” Nature Materials March 02, 2022 (https://doi.org/10/1038/s41563-022-01202-8)
-ProfileProfessor YongKeun ParkBiomedical Optics Laboratory (http://bmol.kaist.ac.kr)Department of PhysicsCollege of Natural SciencesKAIST
2022.03.22 View 10548
Tomographic Measurement of Dielectric Tensors
Dielectric tensor tomography allows the direct measurement of the 3D dielectric tensors of optically anisotropic structures
A research team reported the direct measurement of dielectric tensors of anisotropic structures including the spatial variations of principal refractive indices and directors. The group also demonstrated quantitative tomographic measurements of various nematic liquid-crystal structures and their fast 3D nonequilibrium dynamics using a 3D label-free tomographic method. The method was described in Nature Materials.
Light-matter interactions are described by the dielectric tensor. Despite their importance in basic science and applications, it has not been possible to measure 3D dielectric tensors directly. The main challenge was due to the vectorial nature of light scattering from a 3D anisotropic structure. Previous approaches only addressed 3D anisotropic information indirectly and were limited to two-dimensional, qualitative, strict sample conditions or assumptions.
The research team developed a method enabling the tomographic reconstruction of 3D dielectric tensors without any preparation or assumptions. A sample is illuminated with a laser beam with various angles and circularly polarization states. Then, the light fields scattered from a sample are holographically measured and converted into vectorial diffraction components. Finally, by inversely solving a vectorial wave equation, the 3D dielectric tensor is reconstructed.
Professor YongKeun Park said, “There were a greater number of unknowns in direct measuring than with the conventional approach. We applied our approach to measure additional holographic images by slightly tilting the incident angle.”
He said that the slightly tilted illumination provides an additional orthogonal polarization, which makes the underdetermined problem become the determined problem. “Although scattered fields are dependent on the illumination angle, the Fourier differentiation theorem enables the extraction of the same dielectric tensor for the slightly tilted illumination,” Professor Park added.
His team’s method was validated by reconstructing well-known liquid crystal (LC) structures, including the twisted nematic, hybrid aligned nematic, radial, and bipolar configurations. Furthermore, the research team demonstrated the experimental measurements of the non-equilibrium dynamics of annihilating, nucleating, and merging LC droplets, and the LC polymer network with repeating 3D topological defects.
“This is the first experimental measurement of non-equilibrium dynamics and 3D topological defects in LC structures in a label-free manner. Our method enables the exploration of inaccessible nematic structures and interactions in non-equilibrium dynamics,” first author Dr. Seungwoo Shin explained.
-PublicationSeungwoo Shin, Jonghee Eun, Sang Seok Lee, Changjae Lee, Herve Hugonnet, Dong Ki Yoon, Shin-Hyun Kim, Jongwoo Jeong, YongKeun Park, “Tomographic Measurement ofDielectric Tensors at Optical Frequency,” Nature Materials March 02, 2022 (https://doi.org/10/1038/s41563-022-01202-8)
-ProfileProfessor YongKeun ParkBiomedical Optics Laboratory (http://bmol.kaist.ac.kr)Department of PhysicsCollege of Natural SciencesKAIST
2022.03.22 View 10548 -
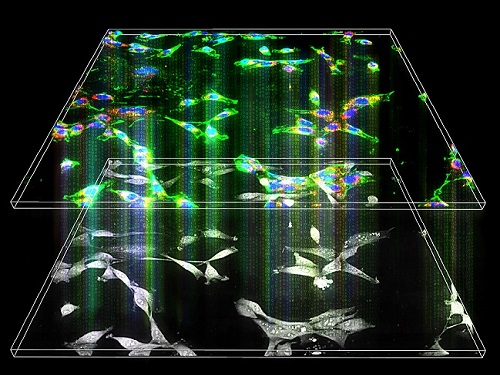 Label-Free Multiplexed Microtomography of Endogenous Subcellular Dynamics Using Deep Learning
AI-based holographic microscopy allows molecular imaging without introducing exogenous labeling agents
A research team upgraded the 3D microtomography observing dynamics of label-free live cells in multiplexed fluorescence imaging. The AI-powered 3D holotomographic microscopy extracts various molecular information from live unlabeled biological cells in real time without exogenous labeling or staining agents.
Professor YongKeum Park’s team and the startup Tomocube encoded 3D refractive index tomograms using the refractive index as a means of measurement. Then they decoded the information with a deep learning-based model that infers multiple 3D fluorescence tomograms from the refractive index measurements of the corresponding subcellular targets, thereby achieving multiplexed micro tomography. This study was reported in Nature Cell Biology online on December 7, 2021.
Fluorescence microscopy is the most widely used optical microscopy technique due to its high biochemical specificity. However, it needs to genetically manipulate or to stain cells with fluorescent labels in order to express fluorescent proteins. These labeling processes inevitably affect the intrinsic physiology of cells. It also has challenges in long-term measuring due to photobleaching and phototoxicity. The overlapped spectra of multiplexed fluorescence signals also hinder the viewing of various structures at the same time. More critically, it took several hours to observe the cells after preparing them.
3D holographic microscopy, also known as holotomography, is providing new ways to quantitatively image live cells without pretreatments such as staining. Holotomography can accurately and quickly measure the morphological and structural information of cells, but only provides limited biochemical and molecular information.
The 'AI microscope' created in this process takes advantage of the features of both holographic microscopy and fluorescence microscopy. That is, a specific image from a fluorescence microscope can be obtained without a fluorescent label. Therefore, the microscope can observe many types of cellular structures in their natural state in 3D and at the same time as fast as one millisecond, and long-term measurements over several days are also possible.
The Tomocube-KAIST team showed that fluorescence images can be directly and precisely predicted from holotomographic images in various cells and conditions. Using the quantitative relationship between the spatial distribution of the refractive index found by AI and the major structures in cells, it was possible to decipher the spatial distribution of the refractive index. And surprisingly, it confirmed that this relationship is constant regardless of cell type.
Professor Park said, “We were able to develop a new concept microscope that combines the advantages of several microscopes with the multidisciplinary research of AI, optics, and biology. It will be immediately applicable for new types of cells not included in the existing data and is expected to be widely applicable for various biological and medical research.”
When comparing the molecular image information extracted by AI with the molecular image information physically obtained by fluorescence staining in 3D space, it showed a 97% or more conformity, which is a level that is difficult to distinguish with the naked eye.
“Compared to the sub-60% accuracy of the fluorescence information extracted from the model developed by the Google AI team, it showed significantly higher performance,” Professor Park added.
This work was supported by the KAIST Up program, the BK21+ program, Tomocube, the National Research Foundation of Korea, and the Ministry of Science and ICT, and the Ministry of Health & Welfare.
-Publication
Hyun-seok Min, Won-Do Heo, YongKeun Park, et al. “Label-free multiplexed microtomography of endogenous subcellular dynamics using generalizable deep learning,” Nature Cell Biology (doi.org/10.1038/s41556-021-00802-x) published online December 07 2021.
-Profile
Professor YongKeun Park
Biomedical Optics Laboratory
Department of Physics
KAIST
2022.02.09 View 13878
Label-Free Multiplexed Microtomography of Endogenous Subcellular Dynamics Using Deep Learning
AI-based holographic microscopy allows molecular imaging without introducing exogenous labeling agents
A research team upgraded the 3D microtomography observing dynamics of label-free live cells in multiplexed fluorescence imaging. The AI-powered 3D holotomographic microscopy extracts various molecular information from live unlabeled biological cells in real time without exogenous labeling or staining agents.
Professor YongKeum Park’s team and the startup Tomocube encoded 3D refractive index tomograms using the refractive index as a means of measurement. Then they decoded the information with a deep learning-based model that infers multiple 3D fluorescence tomograms from the refractive index measurements of the corresponding subcellular targets, thereby achieving multiplexed micro tomography. This study was reported in Nature Cell Biology online on December 7, 2021.
Fluorescence microscopy is the most widely used optical microscopy technique due to its high biochemical specificity. However, it needs to genetically manipulate or to stain cells with fluorescent labels in order to express fluorescent proteins. These labeling processes inevitably affect the intrinsic physiology of cells. It also has challenges in long-term measuring due to photobleaching and phototoxicity. The overlapped spectra of multiplexed fluorescence signals also hinder the viewing of various structures at the same time. More critically, it took several hours to observe the cells after preparing them.
3D holographic microscopy, also known as holotomography, is providing new ways to quantitatively image live cells without pretreatments such as staining. Holotomography can accurately and quickly measure the morphological and structural information of cells, but only provides limited biochemical and molecular information.
The 'AI microscope' created in this process takes advantage of the features of both holographic microscopy and fluorescence microscopy. That is, a specific image from a fluorescence microscope can be obtained without a fluorescent label. Therefore, the microscope can observe many types of cellular structures in their natural state in 3D and at the same time as fast as one millisecond, and long-term measurements over several days are also possible.
The Tomocube-KAIST team showed that fluorescence images can be directly and precisely predicted from holotomographic images in various cells and conditions. Using the quantitative relationship between the spatial distribution of the refractive index found by AI and the major structures in cells, it was possible to decipher the spatial distribution of the refractive index. And surprisingly, it confirmed that this relationship is constant regardless of cell type.
Professor Park said, “We were able to develop a new concept microscope that combines the advantages of several microscopes with the multidisciplinary research of AI, optics, and biology. It will be immediately applicable for new types of cells not included in the existing data and is expected to be widely applicable for various biological and medical research.”
When comparing the molecular image information extracted by AI with the molecular image information physically obtained by fluorescence staining in 3D space, it showed a 97% or more conformity, which is a level that is difficult to distinguish with the naked eye.
“Compared to the sub-60% accuracy of the fluorescence information extracted from the model developed by the Google AI team, it showed significantly higher performance,” Professor Park added.
This work was supported by the KAIST Up program, the BK21+ program, Tomocube, the National Research Foundation of Korea, and the Ministry of Science and ICT, and the Ministry of Health & Welfare.
-Publication
Hyun-seok Min, Won-Do Heo, YongKeun Park, et al. “Label-free multiplexed microtomography of endogenous subcellular dynamics using generalizable deep learning,” Nature Cell Biology (doi.org/10.1038/s41556-021-00802-x) published online December 07 2021.
-Profile
Professor YongKeun Park
Biomedical Optics Laboratory
Department of Physics
KAIST
2022.02.09 View 13878 -
 The Dynamic Tracking of Tissue-Specific Secretory Proteins
Researchers develop a versatile and powerful tool for studying the spatiotemporal dynamics of secretory proteins, a valuable class of biomarkers and therapeutic targets
Researchers have presented a method for profiling tissue-specific secretory proteins in live mice. This method is expected to be applicable to various tissues or disease models for investigating biomarkers or therapeutic targets involved in disease progression. This research was reported in Nature Communications on September 1.
Secretory proteins released into the blood play essential roles in physiological systems. They are core mediators of interorgan communication, while serving as biomarkers and therapeutic targets.
Previous studies have analyzed conditioned media from culture models to identify cell type-specific secretory proteins, but these models often fail to fully recapitulate the intricacies of multi-organ systems and thus do not sufficiently reflect biological realities.
These limitations provided compelling motivation for the research team led by Jae Myoung Suh and his collaborators to develop techniques that could identify and resolve characteristics of tissue-specific secretory proteins along time and space dimensions.
For addressing this gap in the current methodology, the research team utilized proximity-labeling enzymes such as TurboID to label secretory proteins in endoplasmic reticulum lumen using biotin. Thereafter, the biotin-labeled secretory proteins were readily enriched through streptavidin affinity purification and could be identified through mass spectrometry.
To demonstrate its functionality in live mice, research team delivered TurboID to mouse livers via an adenovirus. After administering the biotin, only liver-derived secretory proteins were successfully detected in the plasma of the mice. Interestingly, the pattern of biotin-labeled proteins secreted from the liver was clearly distinctive from those of hepatocyte cell lines.
First author Kwang-eun Kim from the Graduate School of Medical Science and Engineering explained, “The proteins secreted by the liver were significantly different from the results of cell culture models. This data shows the limitations of cell culture models for secretory protein study, and this technique can overcome those limitations. It can be further used to discover biomarkers and therapeutic targets that can more fully reflect the physiological state.”
This work research was supported by the National Research Foundation of Korea, the KAIST Key Research Institutes Project (Interdisciplinary Research Group), and the Institute for Basic Science in Korea.
-PublicationKwang-eun Kim, Isaac Park et al., “Dynamic tracking and identification of tissue-specific secretory proteins in the circulation of live mice,” Nature Communications on Sept.1,
2021(https://doi.org/10.1038/s41467-021-25546-y)
-ProfileProfessor Jae Myoung Suh Integrated Lab of Metabolism, Obesity and Diabetes Researchhttps://imodkaist.wixsite.com/home
Graduate School of Medical Science and Engineering College of Life Science and BioengineeringKAIST
2021.09.14 View 11418
The Dynamic Tracking of Tissue-Specific Secretory Proteins
Researchers develop a versatile and powerful tool for studying the spatiotemporal dynamics of secretory proteins, a valuable class of biomarkers and therapeutic targets
Researchers have presented a method for profiling tissue-specific secretory proteins in live mice. This method is expected to be applicable to various tissues or disease models for investigating biomarkers or therapeutic targets involved in disease progression. This research was reported in Nature Communications on September 1.
Secretory proteins released into the blood play essential roles in physiological systems. They are core mediators of interorgan communication, while serving as biomarkers and therapeutic targets.
Previous studies have analyzed conditioned media from culture models to identify cell type-specific secretory proteins, but these models often fail to fully recapitulate the intricacies of multi-organ systems and thus do not sufficiently reflect biological realities.
These limitations provided compelling motivation for the research team led by Jae Myoung Suh and his collaborators to develop techniques that could identify and resolve characteristics of tissue-specific secretory proteins along time and space dimensions.
For addressing this gap in the current methodology, the research team utilized proximity-labeling enzymes such as TurboID to label secretory proteins in endoplasmic reticulum lumen using biotin. Thereafter, the biotin-labeled secretory proteins were readily enriched through streptavidin affinity purification and could be identified through mass spectrometry.
To demonstrate its functionality in live mice, research team delivered TurboID to mouse livers via an adenovirus. After administering the biotin, only liver-derived secretory proteins were successfully detected in the plasma of the mice. Interestingly, the pattern of biotin-labeled proteins secreted from the liver was clearly distinctive from those of hepatocyte cell lines.
First author Kwang-eun Kim from the Graduate School of Medical Science and Engineering explained, “The proteins secreted by the liver were significantly different from the results of cell culture models. This data shows the limitations of cell culture models for secretory protein study, and this technique can overcome those limitations. It can be further used to discover biomarkers and therapeutic targets that can more fully reflect the physiological state.”
This work research was supported by the National Research Foundation of Korea, the KAIST Key Research Institutes Project (Interdisciplinary Research Group), and the Institute for Basic Science in Korea.
-PublicationKwang-eun Kim, Isaac Park et al., “Dynamic tracking and identification of tissue-specific secretory proteins in the circulation of live mice,” Nature Communications on Sept.1,
2021(https://doi.org/10.1038/s41467-021-25546-y)
-ProfileProfessor Jae Myoung Suh Integrated Lab of Metabolism, Obesity and Diabetes Researchhttps://imodkaist.wixsite.com/home
Graduate School of Medical Science and Engineering College of Life Science and BioengineeringKAIST
2021.09.14 View 11418 -
 New Nanoparticle Drug Combination For Atherosclerosis
Physicochemical cargo-switching nanoparticles (CSNP) designed by KAIST can help significantly reduce cholesterol and macrophage foam cells in arteries, which are the two main triggers for atherosclerotic plaque and inflammation.
The CSNP-based combination drug delivery therapy was proved to exert cholesterol-lowering, anti-inflammatory, and anti-proliferative functions of two common medications for treating and preventing atherosclerosis that are cyclodextrin and statin. Professor Ji-Ho Park and Dr. Heegon Kim from KAIST’s Department of Bio and Brain Engineering said their study has shown great potential for future applications with reduced side effects.
Atherosclerosis is a chronic inflammatory vascular disease that is characterized by the accumulation of cholesterol and cholesterol-loaded macrophage foam cells in the intima. When this atherosclerotic plaque clogs and narrows the artery walls, they restrict blood flow and cause various cardiovascular conditions such as heart attacks and strokes. Heart attacks and strokes are the world’s first and fifth causes of death respectively.
Oral statin administration has been used in clinics as a standard care for atherosclerosis, which is prescribed to lower blood cholesterol and inhibit its accumulation within the plaque. Although statins can effectively prevent the progression of plaque growth, they have only shown modest efficacy in eliminating the already-established plaque. Therefore, patients are required to take statin drugs for the rest of their lives and will always carry the risk of plaque ruptures that can trigger a blood clot.
To address these issues, Professor Park and Dr. Kim exploited another antiatherogenic agent called cyclodextrin. In their paper published in the Journal of Controlled Release on March 10, Professor Park and Dr. Kim reported that the polymeric formulation of cyclodextrin with a diameter of approximately 10 nanometers(nm) can accumulate within the atherosclerotic plaque 14 times more and effectively reduce the plaque even at lower doses, compared to cyclodextrin in a non-polymer structure.
Moreover, although cyclodextrin is known to have a cytotoxic effect on hair cells in the cochlea, which can lead to hearing loss, cyclodextrin polymers developed by Professor Park’s research group exhibited a varying biodistribution profile and did not have this side effect.
In the follow-up study reported in ACS Nano on April 28, the researchers exploited both cyclodextrin and statin and form the cyclodextrin-statin self-assembly drug complex, based on previous findings that each drug can exert local anti-atherosclerosis effect within the plaque. The complex formation processes were optimized to obtain homogeneous and stable nanoparticles with a diameter of about 100 nm for systematic injection.
The therapeutic synergy of cyclodextrin and statin could reportedly enhance plaque-targeted drug delivery and anti-inflammation. Cyclodextrin led to the regression of cholesterol in the established plaque, and the statins were shown to inhibit the proliferation of macrophage foam cells. The study suggested that combination therapy is required to resolve the complex inflammatory cholesterol-rich microenvironment within the plaque.
Professor Park said, “While nanomedicine has been mainly developed for the treatment of cancers, our studies show that nanomedicine can also play a significant role in treating and preventing atherosclerosis, which causes various cardiovascular diseases that are the leading causes of death worldwide.”
This work was supported by KAIST and the National Research Foundation (NRF) of Korea.
Publications:
1. Heegon Kim, Junhee Han, and Ji-Ho Park. (2020) ‘Cyclodextrin polymer improves atherosclerosis therapy and reduces ototoxicity’ Journal of Controlled Release. Volume 319. Page 77-86. Available online at https://doi.org/10.1016/j.jconrel.2019.12.021
2. Kim, H., et al. (2020) ‘Affinity-Driven Design of Cargo-Switching Nanoparticles to Leverage a Cholesterol-Rich Microenvironment for Atherosclerosis Therapy’ ACS Nano. Available online at https://doi.org/10.1021/acsnano.9b08216
Profile: Ji-Ho Park, Ph.D.
Associate Professor
jihopark@kaist.ac.kr
http://openwetware.org/wiki/Park_Lab
Biomaterials Engineering Laboratory (BEL)
Department of Bio and Brain Engineering (BIOENG)
Korea Advanced Institute of Science and Technology (KAIST)
https://www.kaist.ac.kr
Daejeon 34141, Korea
Profile: Heegon Kim, Ph.D.
Postdoctoral Researcher
heegon@kaist.ac.kr
BEL, BIOENG, KAIST
(END)
2020.06.16 View 16500
New Nanoparticle Drug Combination For Atherosclerosis
Physicochemical cargo-switching nanoparticles (CSNP) designed by KAIST can help significantly reduce cholesterol and macrophage foam cells in arteries, which are the two main triggers for atherosclerotic plaque and inflammation.
The CSNP-based combination drug delivery therapy was proved to exert cholesterol-lowering, anti-inflammatory, and anti-proliferative functions of two common medications for treating and preventing atherosclerosis that are cyclodextrin and statin. Professor Ji-Ho Park and Dr. Heegon Kim from KAIST’s Department of Bio and Brain Engineering said their study has shown great potential for future applications with reduced side effects.
Atherosclerosis is a chronic inflammatory vascular disease that is characterized by the accumulation of cholesterol and cholesterol-loaded macrophage foam cells in the intima. When this atherosclerotic plaque clogs and narrows the artery walls, they restrict blood flow and cause various cardiovascular conditions such as heart attacks and strokes. Heart attacks and strokes are the world’s first and fifth causes of death respectively.
Oral statin administration has been used in clinics as a standard care for atherosclerosis, which is prescribed to lower blood cholesterol and inhibit its accumulation within the plaque. Although statins can effectively prevent the progression of plaque growth, they have only shown modest efficacy in eliminating the already-established plaque. Therefore, patients are required to take statin drugs for the rest of their lives and will always carry the risk of plaque ruptures that can trigger a blood clot.
To address these issues, Professor Park and Dr. Kim exploited another antiatherogenic agent called cyclodextrin. In their paper published in the Journal of Controlled Release on March 10, Professor Park and Dr. Kim reported that the polymeric formulation of cyclodextrin with a diameter of approximately 10 nanometers(nm) can accumulate within the atherosclerotic plaque 14 times more and effectively reduce the plaque even at lower doses, compared to cyclodextrin in a non-polymer structure.
Moreover, although cyclodextrin is known to have a cytotoxic effect on hair cells in the cochlea, which can lead to hearing loss, cyclodextrin polymers developed by Professor Park’s research group exhibited a varying biodistribution profile and did not have this side effect.
In the follow-up study reported in ACS Nano on April 28, the researchers exploited both cyclodextrin and statin and form the cyclodextrin-statin self-assembly drug complex, based on previous findings that each drug can exert local anti-atherosclerosis effect within the plaque. The complex formation processes were optimized to obtain homogeneous and stable nanoparticles with a diameter of about 100 nm for systematic injection.
The therapeutic synergy of cyclodextrin and statin could reportedly enhance plaque-targeted drug delivery and anti-inflammation. Cyclodextrin led to the regression of cholesterol in the established plaque, and the statins were shown to inhibit the proliferation of macrophage foam cells. The study suggested that combination therapy is required to resolve the complex inflammatory cholesterol-rich microenvironment within the plaque.
Professor Park said, “While nanomedicine has been mainly developed for the treatment of cancers, our studies show that nanomedicine can also play a significant role in treating and preventing atherosclerosis, which causes various cardiovascular diseases that are the leading causes of death worldwide.”
This work was supported by KAIST and the National Research Foundation (NRF) of Korea.
Publications:
1. Heegon Kim, Junhee Han, and Ji-Ho Park. (2020) ‘Cyclodextrin polymer improves atherosclerosis therapy and reduces ototoxicity’ Journal of Controlled Release. Volume 319. Page 77-86. Available online at https://doi.org/10.1016/j.jconrel.2019.12.021
2. Kim, H., et al. (2020) ‘Affinity-Driven Design of Cargo-Switching Nanoparticles to Leverage a Cholesterol-Rich Microenvironment for Atherosclerosis Therapy’ ACS Nano. Available online at https://doi.org/10.1021/acsnano.9b08216
Profile: Ji-Ho Park, Ph.D.
Associate Professor
jihopark@kaist.ac.kr
http://openwetware.org/wiki/Park_Lab
Biomaterials Engineering Laboratory (BEL)
Department of Bio and Brain Engineering (BIOENG)
Korea Advanced Institute of Science and Technology (KAIST)
https://www.kaist.ac.kr
Daejeon 34141, Korea
Profile: Heegon Kim, Ph.D.
Postdoctoral Researcher
heegon@kaist.ac.kr
BEL, BIOENG, KAIST
(END)
2020.06.16 View 16500 -
 Deep Learning-Powered 'DeepEC' Helps Accurately Understand Enzyme Functions
(Figure: Overall scheme of DeepEC)
A deep learning-powered computational framework, ‘DeepEC,’ will allow the high-quality and high-throughput prediction of enzyme commission numbers, which is essential for the accurate understanding of enzyme functions.
A team of Dr. Jae Yong Ryu, Professor Hyun Uk Kim, and Distinguished Professor Sang Yup Lee at KAIST reported the computational framework powered by deep learning that predicts enzyme commission (EC) numbers with high precision in a high-throughput manner.
DeepEC takes a protein sequence as an input and accurately predicts EC numbers as an output. Enzymes are proteins that catalyze biochemical reactions and EC numbers consisting of four level numbers (i.e., a.b.c.d) indicate biochemical reactions. Thus, the identification of EC numbers is critical for accurately understanding enzyme functions and metabolism.
EC numbers are usually given to a protein sequence encoding an enzyme during a genome annotation procedure. Because of the importance of EC numbers, several EC number prediction tools have been developed, but they have room for further improvement with respect to computation time, precision, coverage, and the total size of the files needed for the EC number prediction.
DeepEC uses three convolutional neural networks (CNNs) as a major engine for the prediction of EC numbers, and also implements homology analysis for EC numbers if the three CNNs do not produce reliable EC numbers for a given protein sequence. DeepEC was developed by using a gold standard dataset covering 1,388,606 protein sequences and 4,669 EC numbers.
In particular, benchmarking studies of DeepEC and five other representative EC number prediction tools showed that DeepEC made the most precise and fastest predictions for EC numbers. DeepEC also required the smallest disk space for implementation, which makes it an ideal third-party software component.
Furthermore, DeepEC was the most sensitive in detecting enzymatic function loss as a result of mutations in domains/binding site residue of protein sequences; in this comparative analysis, all the domains or binding site residue were substituted with L-alanine residue in order to remove the protein function, which is known as the L-alanine scanning method.
This study was published online in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 20, 2019, entitled “Deep learning enables high-quality and high-throughput prediction of enzyme commission numbers.”
“DeepEC can be used as an independent tool and also as a third-party software component in combination with other computational platforms that examine metabolic reactions. DeepEC is freely available online,” said Professor Kim.
Distinguished Professor Lee said, “With DeepEC, it has become possible to process ever-increasing volumes of protein sequence data more efficiently and more accurately.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science and ICT through the National Research Foundation of Korea. This work was also funded by the Bio & Medical Technology Development Program of the National Research Foundation of Korea funded by the Korean government, the Ministry of Science and ICT.
Profile:
-Professor Hyun Uk Kim (ehukim@kaist.ac.kr)
https://sites.google.com/view/ehukim
Department of Chemical and Biomolecular Engineering
-Distinguished Professor Sang Yup Lee (leesy@kaist.ac.kr)
Department of Chemical and Biomolecular Engineering
http://mbel.kaist.ac.kr
2019.07.09 View 41188
Deep Learning-Powered 'DeepEC' Helps Accurately Understand Enzyme Functions
(Figure: Overall scheme of DeepEC)
A deep learning-powered computational framework, ‘DeepEC,’ will allow the high-quality and high-throughput prediction of enzyme commission numbers, which is essential for the accurate understanding of enzyme functions.
A team of Dr. Jae Yong Ryu, Professor Hyun Uk Kim, and Distinguished Professor Sang Yup Lee at KAIST reported the computational framework powered by deep learning that predicts enzyme commission (EC) numbers with high precision in a high-throughput manner.
DeepEC takes a protein sequence as an input and accurately predicts EC numbers as an output. Enzymes are proteins that catalyze biochemical reactions and EC numbers consisting of four level numbers (i.e., a.b.c.d) indicate biochemical reactions. Thus, the identification of EC numbers is critical for accurately understanding enzyme functions and metabolism.
EC numbers are usually given to a protein sequence encoding an enzyme during a genome annotation procedure. Because of the importance of EC numbers, several EC number prediction tools have been developed, but they have room for further improvement with respect to computation time, precision, coverage, and the total size of the files needed for the EC number prediction.
DeepEC uses three convolutional neural networks (CNNs) as a major engine for the prediction of EC numbers, and also implements homology analysis for EC numbers if the three CNNs do not produce reliable EC numbers for a given protein sequence. DeepEC was developed by using a gold standard dataset covering 1,388,606 protein sequences and 4,669 EC numbers.
In particular, benchmarking studies of DeepEC and five other representative EC number prediction tools showed that DeepEC made the most precise and fastest predictions for EC numbers. DeepEC also required the smallest disk space for implementation, which makes it an ideal third-party software component.
Furthermore, DeepEC was the most sensitive in detecting enzymatic function loss as a result of mutations in domains/binding site residue of protein sequences; in this comparative analysis, all the domains or binding site residue were substituted with L-alanine residue in order to remove the protein function, which is known as the L-alanine scanning method.
This study was published online in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 20, 2019, entitled “Deep learning enables high-quality and high-throughput prediction of enzyme commission numbers.”
“DeepEC can be used as an independent tool and also as a third-party software component in combination with other computational platforms that examine metabolic reactions. DeepEC is freely available online,” said Professor Kim.
Distinguished Professor Lee said, “With DeepEC, it has become possible to process ever-increasing volumes of protein sequence data more efficiently and more accurately.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science and ICT through the National Research Foundation of Korea. This work was also funded by the Bio & Medical Technology Development Program of the National Research Foundation of Korea funded by the Korean government, the Ministry of Science and ICT.
Profile:
-Professor Hyun Uk Kim (ehukim@kaist.ac.kr)
https://sites.google.com/view/ehukim
Department of Chemical and Biomolecular Engineering
-Distinguished Professor Sang Yup Lee (leesy@kaist.ac.kr)
Department of Chemical and Biomolecular Engineering
http://mbel.kaist.ac.kr
2019.07.09 View 41188 -
 In Jin Cho Earned the Best Poster Prize at ME Summit 2017
In Jin Cho, a Ph.D. student in the Department of Chemical and Biomolecular Engineering at KAIST received the best poster prize at the International Metabolic Engineering Summit 2017 held on October 24 in Beijing, China.
The International Metabolic Engineering Summit is a global conference where scientists and corporate researchers in the field of metabolic engineering present their latest research outcomes and build networks.
At this year’s summit, about 500 researchers from around the world participated in active academic exchanges, including giving keynote speeches and presenting posters.
During the poster session, the summit selects one person for the KeAi-synthetic and Systems Biotechnology Poster Award, two for Microbial Cell Factories Poster Awards, and three for Biotechnology Journal Poster Awards among the posters presented by graduate students, post-doctoral fellows and researchers. Cho received the KeAi-synthetic and Systems Biotechnology Poster Award. Her winning poster is on the biotransformation of p-xylene to terephthalic acid using engineered Escherichia coli.
Terephthalic acid is generally produced by p-xylene oxidation; however, this process requires a high temperature and pressure as well as a toxic catalyst during the reaction process.
Cho and Ziwei Luo, a Ph.D. student at KAIST, co-conducted the research and developed a successful biological conversion process. Compared to the existing chemical process, it does not require a high temperature and pressure; and it is environmentally friendly with a relatively high conversion rate of approximately 97%.
Cho’s advisor, Distinguished Professor Sang Yup Lee said, “Further research on glucose-derived terephthalic acid will enable us to produce biomass-based eco-friendly terephthalic acid through engineered Escherichia coli.”
2017.10.31 View 12662
In Jin Cho Earned the Best Poster Prize at ME Summit 2017
In Jin Cho, a Ph.D. student in the Department of Chemical and Biomolecular Engineering at KAIST received the best poster prize at the International Metabolic Engineering Summit 2017 held on October 24 in Beijing, China.
The International Metabolic Engineering Summit is a global conference where scientists and corporate researchers in the field of metabolic engineering present their latest research outcomes and build networks.
At this year’s summit, about 500 researchers from around the world participated in active academic exchanges, including giving keynote speeches and presenting posters.
During the poster session, the summit selects one person for the KeAi-synthetic and Systems Biotechnology Poster Award, two for Microbial Cell Factories Poster Awards, and three for Biotechnology Journal Poster Awards among the posters presented by graduate students, post-doctoral fellows and researchers. Cho received the KeAi-synthetic and Systems Biotechnology Poster Award. Her winning poster is on the biotransformation of p-xylene to terephthalic acid using engineered Escherichia coli.
Terephthalic acid is generally produced by p-xylene oxidation; however, this process requires a high temperature and pressure as well as a toxic catalyst during the reaction process.
Cho and Ziwei Luo, a Ph.D. student at KAIST, co-conducted the research and developed a successful biological conversion process. Compared to the existing chemical process, it does not require a high temperature and pressure; and it is environmentally friendly with a relatively high conversion rate of approximately 97%.
Cho’s advisor, Distinguished Professor Sang Yup Lee said, “Further research on glucose-derived terephthalic acid will enable us to produce biomass-based eco-friendly terephthalic acid through engineered Escherichia coli.”
2017.10.31 View 12662 -
 Professor Jae Kyoung Kim Receives the 2017 HSFP Award
The Human Frontier Science Program (HSFP), one of the most competitive research grants in life sciences, has funded researchers worldwide across and beyond the field since 1990. Each year, the program selects a handful of recipients who push the envelope of basic research in biology to bring breakthroughs from novel approaches. Among its 7,000 recipients thus far, 26 scientists have received the Nobel Prize. For that reason, HSFP grants are often referred to as “Nobel Prize Grants.”
Professor Jae Kyoung Kim of the Mathematical Sciences Department at KAIST and his international collaborators, Professor Robert Havekes from the University of Groningen, the Netherlands, Professor Sara Aton from the University of Michigan in Ann Arbor, the United States, and Professor Matias Zurbriggen from the University of Düsseldorf, Germany, won the Young Investigator Grants of the 2017 HSFP.
The 30 winning teams of the 2017 competition (in 9 Young Investigator Grants and 21 Program Grants) went through a rigorous year-long review process from a total of 1,073 applications submitted from more than 60 countries around the world. Each winning team will receive financial support averaging 110,000-125,000 USD per year for three years.
Although Professor Kim was trained as a mathematician, he has extended his research focus into biological sciences and attempted to solve some of the most difficult problems in biology by employing mathematical theories and applications including nonlinear dynamics, stochastic process, singular perturbation, and parameter estimation.
The project that won the Young Investigator Grants was a study on how a molecular circadian clock may affect sleep-regulated neurophysiology in mammals. Physiological and metabolic processes such as sleep, blood pressure, and hormone secretion exhibit circadian rhythms in mammals. Professor Kim used mathematical modeling and analysis to explain that the mammalian circadian clock is a hierarchical system, in which the master clock in the superchiasmatic nucleus, a tiny region in the brain that controls circadian rhythms, functions as a pacemaker and synchronizer of peripheral clocks to generate coherent systematic rhythms throughout the body.
Professor Kim said, “The mechanisms of our neuronal and hormonal activities regulating many of our bodily functions over a 24-hour cycle are not yet fully known. We go to sleep every night, but do not really know how it affects our brain functions. I hope my experience in mathematics, along with insights from biologists, can find meaningful answers to some of today’s puzzling problems in biological sciences, for example, revealing the complexities of our brains and showing how they work.”
“In the meantime, I hope collaborations between the fields of mathematics and biology, as yet a rare phenomenon in the Korean scientific community, will become more popular in the near future.”
Professor Kim received his doctoral degree in Applied and Interdisciplinary Mathematics in 2013 from the University of Michigan and joined KAIST in 2015. He has published numerous articles in reputable science journals such as Science, Molecular Cell, Proceedings of the National Academy of Sciences, and Nature Communications.
Both the Program Grants and Young Investigator Grants support international teams with members from at least two countries for innovative and creative research. This year, the Program Grants were awarded to research topics ranging from the evolution of counting and the role of extracellular vesicles in breast cancer bone metastasis to the examination of obesity from a mechanobiological point of view.
The Young Investigator Grants are limited to teams that established their independent research within the last five years and received their doctoral degrees within the last decade. Besides Professor Kim’s study, such topics as the use of infrasound for navigation by seabirds and protein formation in photochemistry and photophysics were awarded in 2017.
Full lists of the 2017 HFSP winners are available at: http://www.hfsp.org/awardees/newly-awarded.
About the Human Frontier Science Program (HFSP):
The HFSP is a research funding program implemented by the International Human Frontier Science Program (HFSPO) based in Strasbourg, France. It promotes intercontinental collaboration and training in cutting-edge, interdisciplinary research specializing in life sciences. Founded in 1989, the HFSPO consists of the European Union and 14 other countries including the G7 nations and South Korea.
2017.03.21 View 13058
Professor Jae Kyoung Kim Receives the 2017 HSFP Award
The Human Frontier Science Program (HSFP), one of the most competitive research grants in life sciences, has funded researchers worldwide across and beyond the field since 1990. Each year, the program selects a handful of recipients who push the envelope of basic research in biology to bring breakthroughs from novel approaches. Among its 7,000 recipients thus far, 26 scientists have received the Nobel Prize. For that reason, HSFP grants are often referred to as “Nobel Prize Grants.”
Professor Jae Kyoung Kim of the Mathematical Sciences Department at KAIST and his international collaborators, Professor Robert Havekes from the University of Groningen, the Netherlands, Professor Sara Aton from the University of Michigan in Ann Arbor, the United States, and Professor Matias Zurbriggen from the University of Düsseldorf, Germany, won the Young Investigator Grants of the 2017 HSFP.
The 30 winning teams of the 2017 competition (in 9 Young Investigator Grants and 21 Program Grants) went through a rigorous year-long review process from a total of 1,073 applications submitted from more than 60 countries around the world. Each winning team will receive financial support averaging 110,000-125,000 USD per year for three years.
Although Professor Kim was trained as a mathematician, he has extended his research focus into biological sciences and attempted to solve some of the most difficult problems in biology by employing mathematical theories and applications including nonlinear dynamics, stochastic process, singular perturbation, and parameter estimation.
The project that won the Young Investigator Grants was a study on how a molecular circadian clock may affect sleep-regulated neurophysiology in mammals. Physiological and metabolic processes such as sleep, blood pressure, and hormone secretion exhibit circadian rhythms in mammals. Professor Kim used mathematical modeling and analysis to explain that the mammalian circadian clock is a hierarchical system, in which the master clock in the superchiasmatic nucleus, a tiny region in the brain that controls circadian rhythms, functions as a pacemaker and synchronizer of peripheral clocks to generate coherent systematic rhythms throughout the body.
Professor Kim said, “The mechanisms of our neuronal and hormonal activities regulating many of our bodily functions over a 24-hour cycle are not yet fully known. We go to sleep every night, but do not really know how it affects our brain functions. I hope my experience in mathematics, along with insights from biologists, can find meaningful answers to some of today’s puzzling problems in biological sciences, for example, revealing the complexities of our brains and showing how they work.”
“In the meantime, I hope collaborations between the fields of mathematics and biology, as yet a rare phenomenon in the Korean scientific community, will become more popular in the near future.”
Professor Kim received his doctoral degree in Applied and Interdisciplinary Mathematics in 2013 from the University of Michigan and joined KAIST in 2015. He has published numerous articles in reputable science journals such as Science, Molecular Cell, Proceedings of the National Academy of Sciences, and Nature Communications.
Both the Program Grants and Young Investigator Grants support international teams with members from at least two countries for innovative and creative research. This year, the Program Grants were awarded to research topics ranging from the evolution of counting and the role of extracellular vesicles in breast cancer bone metastasis to the examination of obesity from a mechanobiological point of view.
The Young Investigator Grants are limited to teams that established their independent research within the last five years and received their doctoral degrees within the last decade. Besides Professor Kim’s study, such topics as the use of infrasound for navigation by seabirds and protein formation in photochemistry and photophysics were awarded in 2017.
Full lists of the 2017 HFSP winners are available at: http://www.hfsp.org/awardees/newly-awarded.
About the Human Frontier Science Program (HFSP):
The HFSP is a research funding program implemented by the International Human Frontier Science Program (HFSPO) based in Strasbourg, France. It promotes intercontinental collaboration and training in cutting-edge, interdisciplinary research specializing in life sciences. Founded in 1989, the HFSPO consists of the European Union and 14 other countries including the G7 nations and South Korea.
2017.03.21 View 13058