ANS
-
 KAIST Successfully Presents the Future of AI Transformation and Physical AI Strategy at the 1st National Strategic Technology Forum
<(Front row, fourth from the right) President Kwang Hyung Lee of KAIST, (back row, fifth from the right) Forum co-host Representative Hyung-Doo Choi, (back row, sixth from the left) Forum co-host Representative Han-Kyu Kim, along with ruling and opposition party members of the Science, ICT, Broadcasting, and Communications Committee and the Trade, Industry, Energy, SMEs, and Startups Committee, as well as Professors Hoe-Jun Yoo and Jung Kim from KAIST)>
KAIST (President Kwang Hyung Lee) announced on July that it had successfully held the “1st National Strategic Technology Forum” at the National Assembly Members' Office Building that day under the theme “The Future of Artificial Intelligence Transformation (AX): Physical AI.” This bipartisan policy forum aimed to discuss strategies for technology hegemony by leveraging Korea’s strengths in AI semiconductors and manufacturing.
The forum was hosted by KAIST and co-organized by Representative Hyung-Du Choi (People Power Party), the secretary of the National Assembly's Science, ICT, Broadcasting, and Communications Committee, and Representative Han-Kyu Kim (Democratic Party), a member of the Trade, Industry, Energy, SMEs, and Startups Committee. It marks the beginning of a five-part forum series, scheduled monthly through the rest of the year except for October.
The overarching theme, “Artificial Intelligence Transformation (AX),” was designed to address the structural changes reshaping industry, the economy, and society due to the spread of generative AI.
< KAIST President Kwang Hyung Lee delivering his remarks >
The first session focused on “Physical AI,” reflecting how AI innovation—sparked by the proliferation of large language models (LLMs)—is rapidly expanding into the physical realm through ultra-low-power, ultra-lightweight semiconductors. This includes applications in robotics, sensors, and edge devices. Physical AI refers to technologies that interact directly with the real world through AI integration with robotics, autonomous driving, and smart factories. It is drawing attention as a promising next-generation field where Korea can secure a strategic edge, given its strengths in semiconductors and manufacturing.
<Hoi-Jun Yoo, Dean of the KAIST Graduate School of AI Semiconductor>
Hoi-Jun Yoo, Dean of the KAIST Graduate School of AI Semiconductor, gave a presentation titled “The Second AI Innovation Enabled by Ultra-Low-Power AI Semiconductors and Lightweight AI Models,” covering semiconductor trends for implementing Physical AI, academic and industrial strategies for robotics and semiconductors, and Korea’s development direction for “K-Physical AI.”
<Professor Jung Kim, the head of KAIST’s Department of Mechanical and Aerospace Engineering>
Following that, Professor Jung Kim, the head of KAIST’s Department of Mechanical and Aerospace Engineering gave a talk on “Trends in Physical AI and Humanoid Robots,” predicting a new industrial paradigm shaped by AI-robot convergence. He presented global trends, Korea’s development trajectory, and survival strategies for humanoid robots that can supplement or replace human intellectual and physical functions.
During the open discussion that followed, participating lawmakers and experts engaged in in-depth conversations about the need for bipartisan strategies and collaboration.
Representative Hyung-Du Choi (People Power Party) stated, “Through this forum as a platform for public discourse, I will work to ensure that legislation and policy align with the direction of the science and technology field, and that necessary measures are taken promptly to strengthen national competitiveness.”
Representative Han-Kyu Kim (Democratic Party) emphasized, “As strategic planning in science and technology accelerates, it becomes more difficult to coordinate policies involving multiple ministries. Forums like this, which enable ongoing communication among stakeholders, are instrumental in finding effective solutions.”
KAIST President Kwang Hyung Lee remarked, “Although Korea is a latecomer in the generative AI field, we have a unique opportunity to gain strategic superiority in Physical AI, thanks to our technological capabilities in manufacturing, semiconductors, and robotics.” He added, “I hope lawmakers from both the ruling and opposition parties, along with experts, will come together regularly to devise practical policies and contribute to the advancement of Korea’s science and technology.”
<Poster of National Strategic Technology Forum>
This forum series aims to explore policy and institutional solutions to help Korea gain technological leadership in a global context where strategic technologies—such as AI, semiconductors, biotechnology, and energy—directly influence national security and economic sovereignty. Lawmakers from both the Science, ICT, Broadcasting, and Communications Committee and the Trade, Industry, Energy, SMEs, and Startups Committee will continue to participate, fostering bipartisan dialogue. The forums are coordinated by the KAIST Policy Research Institute for National Strategic Technologies.
2025.07.31 View 208
KAIST Successfully Presents the Future of AI Transformation and Physical AI Strategy at the 1st National Strategic Technology Forum
<(Front row, fourth from the right) President Kwang Hyung Lee of KAIST, (back row, fifth from the right) Forum co-host Representative Hyung-Doo Choi, (back row, sixth from the left) Forum co-host Representative Han-Kyu Kim, along with ruling and opposition party members of the Science, ICT, Broadcasting, and Communications Committee and the Trade, Industry, Energy, SMEs, and Startups Committee, as well as Professors Hoe-Jun Yoo and Jung Kim from KAIST)>
KAIST (President Kwang Hyung Lee) announced on July that it had successfully held the “1st National Strategic Technology Forum” at the National Assembly Members' Office Building that day under the theme “The Future of Artificial Intelligence Transformation (AX): Physical AI.” This bipartisan policy forum aimed to discuss strategies for technology hegemony by leveraging Korea’s strengths in AI semiconductors and manufacturing.
The forum was hosted by KAIST and co-organized by Representative Hyung-Du Choi (People Power Party), the secretary of the National Assembly's Science, ICT, Broadcasting, and Communications Committee, and Representative Han-Kyu Kim (Democratic Party), a member of the Trade, Industry, Energy, SMEs, and Startups Committee. It marks the beginning of a five-part forum series, scheduled monthly through the rest of the year except for October.
The overarching theme, “Artificial Intelligence Transformation (AX),” was designed to address the structural changes reshaping industry, the economy, and society due to the spread of generative AI.
< KAIST President Kwang Hyung Lee delivering his remarks >
The first session focused on “Physical AI,” reflecting how AI innovation—sparked by the proliferation of large language models (LLMs)—is rapidly expanding into the physical realm through ultra-low-power, ultra-lightweight semiconductors. This includes applications in robotics, sensors, and edge devices. Physical AI refers to technologies that interact directly with the real world through AI integration with robotics, autonomous driving, and smart factories. It is drawing attention as a promising next-generation field where Korea can secure a strategic edge, given its strengths in semiconductors and manufacturing.
<Hoi-Jun Yoo, Dean of the KAIST Graduate School of AI Semiconductor>
Hoi-Jun Yoo, Dean of the KAIST Graduate School of AI Semiconductor, gave a presentation titled “The Second AI Innovation Enabled by Ultra-Low-Power AI Semiconductors and Lightweight AI Models,” covering semiconductor trends for implementing Physical AI, academic and industrial strategies for robotics and semiconductors, and Korea’s development direction for “K-Physical AI.”
<Professor Jung Kim, the head of KAIST’s Department of Mechanical and Aerospace Engineering>
Following that, Professor Jung Kim, the head of KAIST’s Department of Mechanical and Aerospace Engineering gave a talk on “Trends in Physical AI and Humanoid Robots,” predicting a new industrial paradigm shaped by AI-robot convergence. He presented global trends, Korea’s development trajectory, and survival strategies for humanoid robots that can supplement or replace human intellectual and physical functions.
During the open discussion that followed, participating lawmakers and experts engaged in in-depth conversations about the need for bipartisan strategies and collaboration.
Representative Hyung-Du Choi (People Power Party) stated, “Through this forum as a platform for public discourse, I will work to ensure that legislation and policy align with the direction of the science and technology field, and that necessary measures are taken promptly to strengthen national competitiveness.”
Representative Han-Kyu Kim (Democratic Party) emphasized, “As strategic planning in science and technology accelerates, it becomes more difficult to coordinate policies involving multiple ministries. Forums like this, which enable ongoing communication among stakeholders, are instrumental in finding effective solutions.”
KAIST President Kwang Hyung Lee remarked, “Although Korea is a latecomer in the generative AI field, we have a unique opportunity to gain strategic superiority in Physical AI, thanks to our technological capabilities in manufacturing, semiconductors, and robotics.” He added, “I hope lawmakers from both the ruling and opposition parties, along with experts, will come together regularly to devise practical policies and contribute to the advancement of Korea’s science and technology.”
<Poster of National Strategic Technology Forum>
This forum series aims to explore policy and institutional solutions to help Korea gain technological leadership in a global context where strategic technologies—such as AI, semiconductors, biotechnology, and energy—directly influence national security and economic sovereignty. Lawmakers from both the Science, ICT, Broadcasting, and Communications Committee and the Trade, Industry, Energy, SMEs, and Startups Committee will continue to participate, fostering bipartisan dialogue. The forums are coordinated by the KAIST Policy Research Institute for National Strategic Technologies.
2025.07.31 View 208 -
 KAIST School of Transdisciplinary Studies Is Driving Innovation in Korean Education
<(From Left) Professor Jaeseung Jeong, haed of the School of Transdiciplinary Studies, Dr, Albert Chau, Vice President of Hong Kong Baptist University>
KAIST (President Kwang Hyung Lee) announced on the 24th of July that its School of Transdisciplinary Studies has been consistently showcasing the results of its experiments and practices for educational innovation both domestically and abroad.
On June 27, Professor Jaeseung Jeong, head of the School of Transdisciplinary Studies, was invited to speak at the “Pacific Asia Summit on Transdisciplinary Education 2025 (PASTE 2025)” held at Hong Kong Baptist University. He presented the Korean model of transdisciplinary education under the title “The Philosophy and Achievements of the KAIST School of Transdisciplinary Studies.”
In his talk, Professor Jeong pointed out the limitations of conventional education systems that rely on answer-centered evaluation, perfectionism, and competitiveness, claiming that they hinder creativity and integrative thinking. He then introduced the philosophy and operational practices of the School of Transdisciplinary Studies, which was established in 2019 to overcome these issues.
Professor Jeong outlined five key principles that define the school's educational philosophy: ①a broad and integrative academic foundation, ②student-driven and customized education, ③creativity and execution, ④a sense of social responsibility and global citizenship, and ⑤learning driven by intrinsic motivation and curiosity. He explained that students are admitted without a declared major, allowed to design their own learning plans, and evaluated under a P/NR system* that focuses on growth rather than competition.
*P/NR system: A non-competitive grading system led by KAIST’s School of Transdisciplinary Studies. Instead of traditional letter grades (A/B/C/Fail), students receive Pass (P) or No Record (NR), with the latter not appearing as a failure and not affecting GPA.
Professor Jeong emphasized, “This experiment at KAIST represents a new educational paradigm that values questions over knowledge, culture over structure, and inquiry over competition. Students are bridging academic learning and real-world practice by addressing societal challenges through technology, which could lead to a fundamental shift in global higher education.”
His presentation provided an opportunity to spotlight how KAIST’s experimental approach to nurturing transdisciplinary talent is pointing to new directions for the global education community beyond Korea.
< Hyungjoon Jang, a student at the School of Transdisciplinary Studies>
The achievements of KAIST’s transdisciplinary education model are also reflected in students’ academic accomplishments. Hyungjoon Jang, a student at the School of Transdisciplinary Studies, participated in a collaborative study led by his mentor, Professor Jaekyung Kim in the Department of Mathematical Sciences, along with researchers from Chungnam National University and the Institute for Basic Science (IBS). Their groundbreaking analytical method enables the accurate estimation of inhibition constants using only a single inhibitor concentration. The paper was published in the prestigious journal Nature Communications in June, with Jang listed as co–first author.
Jang played a leading role throughout the research process by developing the experimental methodology, creating a software package to support the method, drafting the manuscript, and engaging in peer review. He also effectively communicated mathematical and statistical models to pharmaceutical experts by mastering presentation techniques and visual explanation strategies, thereby setting a strong example of interdisciplinary collaboration.
He emphasized that “the School of Transdisciplinary Studies’ mentor system allowed regular research feedback and the systematic acquisition of essential knowledge and analytical skills through courses in biochemistry and computational neuroscience.”
This example demonstrates how undergraduate students at the School of Transdisciplinary Studies can take leading roles in cutting-edge interdisciplinary research.
The school’s educational philosophy is also reflected in students’ practical actions. Inseo Jeong, a current student and founder of the startup MPAge Inc., made a meaningful donation to help establish a creative makerspace in the school.
<Inseo Jeong, founder of MPAG>
Inseo Jeong explained that the decision was made to express gratitude for the knowledge gained and the mentorship received from professors, saying that at the School of Transdisciplinary Studies, she learned not only how to solve problems with technology but also how to view society, and that learning has helped her grow. She added, “The deep understanding of humanity and the world emphasized by Professor Jaeseung Jeong will be a great asset not only to entrepreneurs but to all students pursuing diverse paths,” expressing support for her fellow students.
Inseo Jeong collaborated for over two years with Professor Hyunwook Ka of the School of Transdisciplinary Studies on software research for individuals with hearing impairments. After numerous algorithm designs and experimental iterations, their work, which considered the social scalability of technology, was presented at the world-renowned CSUN Assistive Technology Conference held at California State University, Northridge. The project has filed for a patent under KAIST’s name.
※ Presentation title: Evidence-Based Adaptive Transcription for Sign Language Users
KAIST is now working to complete the makerspace on the third floor of the Administrative Annex (N2) in Room 314 with a size of approximately 33 m2 during the summer. The makerspace is expected to serve as a hands-on, integrative learning environment where various ideas can be realized and implemented, playing a key role in fostering students’ creative problem-solving and integrative thinking skills.
KAIST President Kwang Hyung Lee stated, “The School of Transdisciplinary Studies is both an experimental ground and a practical field for overcoming the limitations of traditional education and nurturing global talents with creative problem-solving skills and integrative thinking, which are essential for the future.” He added, “KAIST will continue to lead efforts to cultivate question-asking, inquiry-driven, transdisciplinary talents and propose new paradigms for education and research.”
2025.07.24 View 350
KAIST School of Transdisciplinary Studies Is Driving Innovation in Korean Education
<(From Left) Professor Jaeseung Jeong, haed of the School of Transdiciplinary Studies, Dr, Albert Chau, Vice President of Hong Kong Baptist University>
KAIST (President Kwang Hyung Lee) announced on the 24th of July that its School of Transdisciplinary Studies has been consistently showcasing the results of its experiments and practices for educational innovation both domestically and abroad.
On June 27, Professor Jaeseung Jeong, head of the School of Transdisciplinary Studies, was invited to speak at the “Pacific Asia Summit on Transdisciplinary Education 2025 (PASTE 2025)” held at Hong Kong Baptist University. He presented the Korean model of transdisciplinary education under the title “The Philosophy and Achievements of the KAIST School of Transdisciplinary Studies.”
In his talk, Professor Jeong pointed out the limitations of conventional education systems that rely on answer-centered evaluation, perfectionism, and competitiveness, claiming that they hinder creativity and integrative thinking. He then introduced the philosophy and operational practices of the School of Transdisciplinary Studies, which was established in 2019 to overcome these issues.
Professor Jeong outlined five key principles that define the school's educational philosophy: ①a broad and integrative academic foundation, ②student-driven and customized education, ③creativity and execution, ④a sense of social responsibility and global citizenship, and ⑤learning driven by intrinsic motivation and curiosity. He explained that students are admitted without a declared major, allowed to design their own learning plans, and evaluated under a P/NR system* that focuses on growth rather than competition.
*P/NR system: A non-competitive grading system led by KAIST’s School of Transdisciplinary Studies. Instead of traditional letter grades (A/B/C/Fail), students receive Pass (P) or No Record (NR), with the latter not appearing as a failure and not affecting GPA.
Professor Jeong emphasized, “This experiment at KAIST represents a new educational paradigm that values questions over knowledge, culture over structure, and inquiry over competition. Students are bridging academic learning and real-world practice by addressing societal challenges through technology, which could lead to a fundamental shift in global higher education.”
His presentation provided an opportunity to spotlight how KAIST’s experimental approach to nurturing transdisciplinary talent is pointing to new directions for the global education community beyond Korea.
< Hyungjoon Jang, a student at the School of Transdisciplinary Studies>
The achievements of KAIST’s transdisciplinary education model are also reflected in students’ academic accomplishments. Hyungjoon Jang, a student at the School of Transdisciplinary Studies, participated in a collaborative study led by his mentor, Professor Jaekyung Kim in the Department of Mathematical Sciences, along with researchers from Chungnam National University and the Institute for Basic Science (IBS). Their groundbreaking analytical method enables the accurate estimation of inhibition constants using only a single inhibitor concentration. The paper was published in the prestigious journal Nature Communications in June, with Jang listed as co–first author.
Jang played a leading role throughout the research process by developing the experimental methodology, creating a software package to support the method, drafting the manuscript, and engaging in peer review. He also effectively communicated mathematical and statistical models to pharmaceutical experts by mastering presentation techniques and visual explanation strategies, thereby setting a strong example of interdisciplinary collaboration.
He emphasized that “the School of Transdisciplinary Studies’ mentor system allowed regular research feedback and the systematic acquisition of essential knowledge and analytical skills through courses in biochemistry and computational neuroscience.”
This example demonstrates how undergraduate students at the School of Transdisciplinary Studies can take leading roles in cutting-edge interdisciplinary research.
The school’s educational philosophy is also reflected in students’ practical actions. Inseo Jeong, a current student and founder of the startup MPAge Inc., made a meaningful donation to help establish a creative makerspace in the school.
<Inseo Jeong, founder of MPAG>
Inseo Jeong explained that the decision was made to express gratitude for the knowledge gained and the mentorship received from professors, saying that at the School of Transdisciplinary Studies, she learned not only how to solve problems with technology but also how to view society, and that learning has helped her grow. She added, “The deep understanding of humanity and the world emphasized by Professor Jaeseung Jeong will be a great asset not only to entrepreneurs but to all students pursuing diverse paths,” expressing support for her fellow students.
Inseo Jeong collaborated for over two years with Professor Hyunwook Ka of the School of Transdisciplinary Studies on software research for individuals with hearing impairments. After numerous algorithm designs and experimental iterations, their work, which considered the social scalability of technology, was presented at the world-renowned CSUN Assistive Technology Conference held at California State University, Northridge. The project has filed for a patent under KAIST’s name.
※ Presentation title: Evidence-Based Adaptive Transcription for Sign Language Users
KAIST is now working to complete the makerspace on the third floor of the Administrative Annex (N2) in Room 314 with a size of approximately 33 m2 during the summer. The makerspace is expected to serve as a hands-on, integrative learning environment where various ideas can be realized and implemented, playing a key role in fostering students’ creative problem-solving and integrative thinking skills.
KAIST President Kwang Hyung Lee stated, “The School of Transdisciplinary Studies is both an experimental ground and a practical field for overcoming the limitations of traditional education and nurturing global talents with creative problem-solving skills and integrative thinking, which are essential for the future.” He added, “KAIST will continue to lead efforts to cultivate question-asking, inquiry-driven, transdisciplinary talents and propose new paradigms for education and research.”
2025.07.24 View 350 -
 KAIST Kicks Off the Expansion of its Creative Learning Building, a 50th Anniversary Donation Landmark
KAIST announced on July 10th that it held a groundbreaking ceremony on July 9th for the expansion of its Creative Learning Building. This project, which celebrates the university's 50th anniversary, will become a significant donation-funded landmark and marks the official start of its construction.
<(From left) President Kwang Hyung Lee, Former President Sung-Chul Shin>
The groundbreaking ceremony was attended by key donors who graced the occasion, including KAIST President Kwang Hyung Lee, former President Sung-Chul Shin, Alumni Association President Yoon-Tae Lee, as well as parents and faculty member.
The Creative Learning Building serves as a primary space where KAIST undergraduate and graduate students attend lectures, functioning as a central hub for a variety of classes and talks. It also houses student support departments, including the Student Affairs Office, establishing itself as a student-centric complex that integrates educational, counseling, and welfare functions.
This expansion is more than just an increase in educational facilities; it's being developed as a "donation landmark" embodying KAIST's identity and future vision. Designed with a focus on creative convergence education, this project aims to create a new educational hub that organically combines education, exchange, and welfare functions
The campaign included over 230 participants, including KAIST alumni Byung-gyu Chang, Chairman of Krafton, former Alumni Association President Ki-chul Cha, Dr. Kun-mo Chung (former Minister of Science and Technology), as well as faculty members, parents, and current students. They collectively raised 6.5 billion KRW in donations. The total cost for this expansion project is 9 billion KRW, encompassing a gross floor area of 3,222.92㎡ across five above-ground floors, with completion targeted for September 2026.
2025.07.10 View 504
KAIST Kicks Off the Expansion of its Creative Learning Building, a 50th Anniversary Donation Landmark
KAIST announced on July 10th that it held a groundbreaking ceremony on July 9th for the expansion of its Creative Learning Building. This project, which celebrates the university's 50th anniversary, will become a significant donation-funded landmark and marks the official start of its construction.
<(From left) President Kwang Hyung Lee, Former President Sung-Chul Shin>
The groundbreaking ceremony was attended by key donors who graced the occasion, including KAIST President Kwang Hyung Lee, former President Sung-Chul Shin, Alumni Association President Yoon-Tae Lee, as well as parents and faculty member.
The Creative Learning Building serves as a primary space where KAIST undergraduate and graduate students attend lectures, functioning as a central hub for a variety of classes and talks. It also houses student support departments, including the Student Affairs Office, establishing itself as a student-centric complex that integrates educational, counseling, and welfare functions.
This expansion is more than just an increase in educational facilities; it's being developed as a "donation landmark" embodying KAIST's identity and future vision. Designed with a focus on creative convergence education, this project aims to create a new educational hub that organically combines education, exchange, and welfare functions
The campaign included over 230 participants, including KAIST alumni Byung-gyu Chang, Chairman of Krafton, former Alumni Association President Ki-chul Cha, Dr. Kun-mo Chung (former Minister of Science and Technology), as well as faculty members, parents, and current students. They collectively raised 6.5 billion KRW in donations. The total cost for this expansion project is 9 billion KRW, encompassing a gross floor area of 3,222.92㎡ across five above-ground floors, with completion targeted for September 2026.
2025.07.10 View 504 -
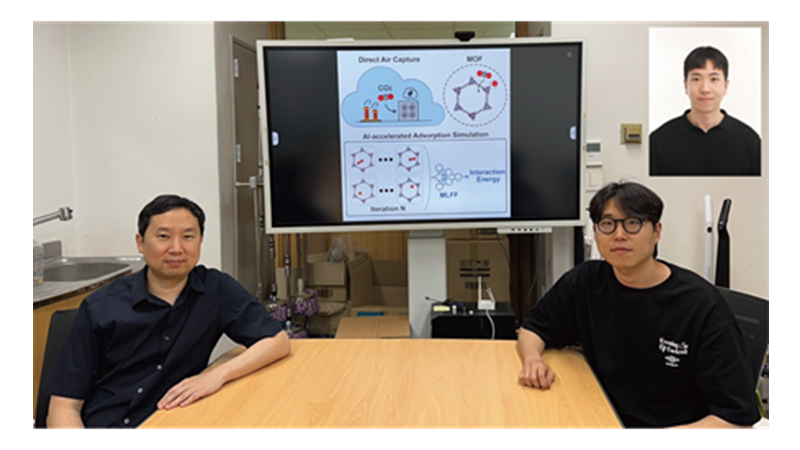 KAIST Develops AI to Easily Find Promising Materials That Capture Only CO₂
< Photo 1. (From left) Professor Jihan Kim, Ph.D. candidate Yunsung Lim and Dr. Hyunsoo Park of the Department of Chemical and Biomolecular Engineering >
In order to help prevent the climate crisis, actively reducing already-emitted CO₂ is essential. Accordingly, direct air capture (DAC) — a technology that directly extracts only CO₂ from the air — is gaining attention. However, effectively capturing pure CO₂ is not easy due to water vapor (H₂O) present in the air. KAIST researchers have successfully used AI-driven machine learning techniques to identify the most promising CO₂-capturing materials among metal-organic frameworks (MOFs), a key class of materials studied for this technology.
KAIST (President Kwang Hyung Lee) announced on the 29th of June that a research team led by Professor Jihan Kim from the Department of Chemical and Biomolecular Engineering, in collaboration with a team at Imperial College London, has developed a machine-learning-based simulation method that can quickly and accurately screen MOFs best suited for atmospheric CO₂ capture.
< Figure 1. Concept diagram of Direct Air Capture (DAC) technology and carbon capture using Metal-Organic Frameworks (MOFs). MOFs are promising porous materials capable of capturing carbon dioxide from the atmosphere, drawing attention as a core material for DAC technology. >
To overcome the difficulty of discovering high-performance materials due to the complexity of structures and the limitations of predicting intermolecular interactions, the research team developed a machine learning force field (MLFF) capable of precisely predicting the interactions between CO₂, water (H₂O), and MOFs. This new method enables calculations of MOF adsorption properties with quantum-mechanics-level accuracy at vastly faster speeds than before.
Using this system, the team screened over 8,000 experimentally synthesized MOF structures, identifying more than 100 promising candidates for CO₂ capture. Notably, this included new candidates that had not been uncovered by traditional force-field-based simulations. The team also analyzed the relationships between MOF chemical structure and adsorption performance, proposing seven key chemical features that will help in designing new materials for DAC.
< Figure 2. Concept diagram of adsorption simulation using Machine Learning Force Field (MLFF). The developed MLFF is applicable to various MOF structures and allows for precise calculation of adsorption properties by predicting interaction energies during repetitive Widom insertion simulations. It is characterized by simultaneously achieving high accuracy and low computational cost compared to conventional classical force fields. >
This research is recognized as a significant advance in the DAC field, greatly enhancing materials design and simulation by precisely predicting MOF-CO₂ and MOF-H₂O interactions.
The results of this research, with Ph.D. candidate Yunsung Lim and Dr. Hyunsoo Park of KAIST as co-first authors, were published in the international academic journal Matter on June 12.
※Paper Title: Accelerating CO₂ direct air capture screening for metal–organic frameworks with a transferable machine learning force field
※DOI: 10.1016/j.matt.2025.102203
This research was supported by the Saudi Aramco-KAIST CO₂ Management Center and the Ministry of Science and ICT's Global C.L.E.A.N. Project.
2025.06.29 View 1419
KAIST Develops AI to Easily Find Promising Materials That Capture Only CO₂
< Photo 1. (From left) Professor Jihan Kim, Ph.D. candidate Yunsung Lim and Dr. Hyunsoo Park of the Department of Chemical and Biomolecular Engineering >
In order to help prevent the climate crisis, actively reducing already-emitted CO₂ is essential. Accordingly, direct air capture (DAC) — a technology that directly extracts only CO₂ from the air — is gaining attention. However, effectively capturing pure CO₂ is not easy due to water vapor (H₂O) present in the air. KAIST researchers have successfully used AI-driven machine learning techniques to identify the most promising CO₂-capturing materials among metal-organic frameworks (MOFs), a key class of materials studied for this technology.
KAIST (President Kwang Hyung Lee) announced on the 29th of June that a research team led by Professor Jihan Kim from the Department of Chemical and Biomolecular Engineering, in collaboration with a team at Imperial College London, has developed a machine-learning-based simulation method that can quickly and accurately screen MOFs best suited for atmospheric CO₂ capture.
< Figure 1. Concept diagram of Direct Air Capture (DAC) technology and carbon capture using Metal-Organic Frameworks (MOFs). MOFs are promising porous materials capable of capturing carbon dioxide from the atmosphere, drawing attention as a core material for DAC technology. >
To overcome the difficulty of discovering high-performance materials due to the complexity of structures and the limitations of predicting intermolecular interactions, the research team developed a machine learning force field (MLFF) capable of precisely predicting the interactions between CO₂, water (H₂O), and MOFs. This new method enables calculations of MOF adsorption properties with quantum-mechanics-level accuracy at vastly faster speeds than before.
Using this system, the team screened over 8,000 experimentally synthesized MOF structures, identifying more than 100 promising candidates for CO₂ capture. Notably, this included new candidates that had not been uncovered by traditional force-field-based simulations. The team also analyzed the relationships between MOF chemical structure and adsorption performance, proposing seven key chemical features that will help in designing new materials for DAC.
< Figure 2. Concept diagram of adsorption simulation using Machine Learning Force Field (MLFF). The developed MLFF is applicable to various MOF structures and allows for precise calculation of adsorption properties by predicting interaction energies during repetitive Widom insertion simulations. It is characterized by simultaneously achieving high accuracy and low computational cost compared to conventional classical force fields. >
This research is recognized as a significant advance in the DAC field, greatly enhancing materials design and simulation by precisely predicting MOF-CO₂ and MOF-H₂O interactions.
The results of this research, with Ph.D. candidate Yunsung Lim and Dr. Hyunsoo Park of KAIST as co-first authors, were published in the international academic journal Matter on June 12.
※Paper Title: Accelerating CO₂ direct air capture screening for metal–organic frameworks with a transferable machine learning force field
※DOI: 10.1016/j.matt.2025.102203
This research was supported by the Saudi Aramco-KAIST CO₂ Management Center and the Ministry of Science and ICT's Global C.L.E.A.N. Project.
2025.06.29 View 1419 -
 KAIST Predicts Diseases by Early Detection of Aging Signals in Liver Tissue
- KAIST-KRIBB Develops ‘FiNi-seq’ Technology to Capture Characteristics of Fibrotic Microenvironments Accumulated in Liver Tissue and Dynamic Changes of Early Aging Cells
- Elucidation of the Spatial Ecosystem of Aged Liver Tissue, where Reprogramming of Senescent Cells and Immune Exhaustion Progresses, at the Single-Cell Genome and Epigenome Levels
< (From left) Professor Jong-Eun Park of KAIST Graduate School of Medical Science and Engineering (GSMSE), Dr. Chuna Kim of KRIBB, Dr. Kwon Yong Tak of KAIST GSMSE, Ph.D. Candidate Juyeon Kim of KRIBB, Ph.D. Candidate Myungsun Park of KAIST GSMSE >
Aging and chronic diseases involve the gradual accumulation of subtle tissue changes over a long period. Therefore, there are still limitations in quantitatively understanding these changes within organs and linking them to early signs of disease onset. In response, Korean researchers have successfully developed a platform technology that accurately captures localized changes that first occur within tissue, significantly aiding in faster disease discovery and prediction, and in setting personalized treatment targets.
KAIST (President Kwang Hyung Lee) announced on June 12th that a joint research team led by Professor Jong-Eun Park of the Graduate School of Medical Science and Engineering at KAIST and Dr. Chuna Kim of the Aging Convergence Research Center at the Korea Research Institute of Bioscience and Biotechnology (KRIBB, President Seok-Yoon Kwon) has developed ‘FiNi-seq (Fibrotic Niche enrichment sequencing)’ technology. This technology captures fibrotic microenvironments locally occurring in aged liver tissue and enables precise analysis at the single-cell transcriptome level*.
*Single-cell transcriptome analysis: A method to measure how actively each cell uses which genes, allowing identification and function of individual diseased cells.
The researchers developed a method to selectively enrich early aging microenvironments where regeneration is delayed and fibrosis accumulates, by physically selecting regions with high tissue degradation resistance in aged liver tissue.
In this process, high-resolution identification of fibrosis-related endothelial cells, fibroblasts interacting with the immune system, and immune-exhausted cells such as PD-1 highly expressing CD8 T cells, which were difficult to capture with existing single-cell analysis technologies, was possible.
In particular, the research team confirmed through ‘FiNi-seq’ technology that specific cells observed in fibrotic areas within aged liver tissue secondarily age the surrounding environment through secreted factors, and that this leads to the expansion of the aged environment.
Furthermore, they also elucidated the mechanism by which endothelial cells lose their tissue-specific identity and induce innate immune responses, promoting immune cell infiltration. Through spatial transcriptome analysis, the spatial distribution of fibroblasts interacting with immune cells was quantified, revealing their involvement in tissue regeneration, induction of inflammatory responses, and progression to chronic fibrosis.
The research team performed integrated analysis of multi-omics\* data to obtain transcriptome and epigenome information, precisely interpreting the microenvironment of aged liver tissue and its spatial heterogeneity, and confirming how these changes are connected to the intrahepatic vascular structure.
*Multi-omics: An integrated analysis method for various biological information within an organism, such as genes, proteins, metabolites, and cell information.
The newly developed ‘FiNi-seq’ technology is expected to be a useful platform for high-resolution capture of pathophysiological signals in most chronic liver diseases, including the aging process that causes fibrosis.
< Figure 1. Isolation of fibrotic regions from aged liver tissue, followed by single-cell transcriptome analysis and validation in a fibrosis model. >
The first author, Dr. Kwon Yong Tak of KAIST Graduate School of Medical Science and Engineering (GSMSE), a hepatologist at Seoul St. Mary's Hospital, designed this study to lay the groundwork for early diagnosis and treatment of fibrosis progression, the most important clinical prognostic indicator in chronic liver disease, while pursuing his Ph.D. at KAIST KAIST GSMSE with support from the physician-scientist training program. Co-first author Myungsun Park, a Ph.D. candidate at KAIST KAIST GSMSE, was responsible for the technical implementation of FiNi-seq technology, and Juyeon Kim, a Ph.D. candidate at KRIBB's Aging Convergence Research Center, was responsible for imaging analysis of aged tissue, playing a key role in the research.
Dr. Chuna Kim of KRIBB stated, “Through this study, we were able to precisely elucidate the cellular composition and spatial characteristics of the fibrotic microenvironment observed in aged liver tissue at the single-cell level.”
< Figure 2. Spatially defined stepwise progression patterns of aging-related regions within the liver and identification of regulatory factors inducing them. >
Professor Jong-Eun Park of the Graduate School of Medical Science and Engineering said, “As an analytical technology that can capture subtle changes occurring in the early stages of aging and chronic diseases, it is expected to play a significant role in finding effective treatment targets in the future. Also, we plan to expand this research to chronic diseases in other organs such as the lungs and kidneys, as well as various liver disease models.”
This research was published in the international journal ‘Nature Aging’ on May 5, 2025, with Dr. Kwon Yong Tak of KAIST KAIST GSMSE, Ph.D. Candidate Juyeon Kim of KRIBB, and Ph.D. Candidate Myungsun Park of KAIST as co-first authors.
*Paper Title: Quasi-spatial single-cell transcriptome based on physical tissue properties defines early aging associated niche in liver
*DOI: https://doi.org/10.1038/s43587-025-00857-7
This research was supported by several domestic institutions, including the National Research Foundation of Korea, the Korea Health Industry Development Institute (KHIDI), the Korea Research Institute of Bioscience and Biotechnology (KRIBB), KIST, POSCO Science Fellowship, and the Convergence Medical Scientist Training Program.
2025.06.12 View 2359
KAIST Predicts Diseases by Early Detection of Aging Signals in Liver Tissue
- KAIST-KRIBB Develops ‘FiNi-seq’ Technology to Capture Characteristics of Fibrotic Microenvironments Accumulated in Liver Tissue and Dynamic Changes of Early Aging Cells
- Elucidation of the Spatial Ecosystem of Aged Liver Tissue, where Reprogramming of Senescent Cells and Immune Exhaustion Progresses, at the Single-Cell Genome and Epigenome Levels
< (From left) Professor Jong-Eun Park of KAIST Graduate School of Medical Science and Engineering (GSMSE), Dr. Chuna Kim of KRIBB, Dr. Kwon Yong Tak of KAIST GSMSE, Ph.D. Candidate Juyeon Kim of KRIBB, Ph.D. Candidate Myungsun Park of KAIST GSMSE >
Aging and chronic diseases involve the gradual accumulation of subtle tissue changes over a long period. Therefore, there are still limitations in quantitatively understanding these changes within organs and linking them to early signs of disease onset. In response, Korean researchers have successfully developed a platform technology that accurately captures localized changes that first occur within tissue, significantly aiding in faster disease discovery and prediction, and in setting personalized treatment targets.
KAIST (President Kwang Hyung Lee) announced on June 12th that a joint research team led by Professor Jong-Eun Park of the Graduate School of Medical Science and Engineering at KAIST and Dr. Chuna Kim of the Aging Convergence Research Center at the Korea Research Institute of Bioscience and Biotechnology (KRIBB, President Seok-Yoon Kwon) has developed ‘FiNi-seq (Fibrotic Niche enrichment sequencing)’ technology. This technology captures fibrotic microenvironments locally occurring in aged liver tissue and enables precise analysis at the single-cell transcriptome level*.
*Single-cell transcriptome analysis: A method to measure how actively each cell uses which genes, allowing identification and function of individual diseased cells.
The researchers developed a method to selectively enrich early aging microenvironments where regeneration is delayed and fibrosis accumulates, by physically selecting regions with high tissue degradation resistance in aged liver tissue.
In this process, high-resolution identification of fibrosis-related endothelial cells, fibroblasts interacting with the immune system, and immune-exhausted cells such as PD-1 highly expressing CD8 T cells, which were difficult to capture with existing single-cell analysis technologies, was possible.
In particular, the research team confirmed through ‘FiNi-seq’ technology that specific cells observed in fibrotic areas within aged liver tissue secondarily age the surrounding environment through secreted factors, and that this leads to the expansion of the aged environment.
Furthermore, they also elucidated the mechanism by which endothelial cells lose their tissue-specific identity and induce innate immune responses, promoting immune cell infiltration. Through spatial transcriptome analysis, the spatial distribution of fibroblasts interacting with immune cells was quantified, revealing their involvement in tissue regeneration, induction of inflammatory responses, and progression to chronic fibrosis.
The research team performed integrated analysis of multi-omics\* data to obtain transcriptome and epigenome information, precisely interpreting the microenvironment of aged liver tissue and its spatial heterogeneity, and confirming how these changes are connected to the intrahepatic vascular structure.
*Multi-omics: An integrated analysis method for various biological information within an organism, such as genes, proteins, metabolites, and cell information.
The newly developed ‘FiNi-seq’ technology is expected to be a useful platform for high-resolution capture of pathophysiological signals in most chronic liver diseases, including the aging process that causes fibrosis.
< Figure 1. Isolation of fibrotic regions from aged liver tissue, followed by single-cell transcriptome analysis and validation in a fibrosis model. >
The first author, Dr. Kwon Yong Tak of KAIST Graduate School of Medical Science and Engineering (GSMSE), a hepatologist at Seoul St. Mary's Hospital, designed this study to lay the groundwork for early diagnosis and treatment of fibrosis progression, the most important clinical prognostic indicator in chronic liver disease, while pursuing his Ph.D. at KAIST KAIST GSMSE with support from the physician-scientist training program. Co-first author Myungsun Park, a Ph.D. candidate at KAIST KAIST GSMSE, was responsible for the technical implementation of FiNi-seq technology, and Juyeon Kim, a Ph.D. candidate at KRIBB's Aging Convergence Research Center, was responsible for imaging analysis of aged tissue, playing a key role in the research.
Dr. Chuna Kim of KRIBB stated, “Through this study, we were able to precisely elucidate the cellular composition and spatial characteristics of the fibrotic microenvironment observed in aged liver tissue at the single-cell level.”
< Figure 2. Spatially defined stepwise progression patterns of aging-related regions within the liver and identification of regulatory factors inducing them. >
Professor Jong-Eun Park of the Graduate School of Medical Science and Engineering said, “As an analytical technology that can capture subtle changes occurring in the early stages of aging and chronic diseases, it is expected to play a significant role in finding effective treatment targets in the future. Also, we plan to expand this research to chronic diseases in other organs such as the lungs and kidneys, as well as various liver disease models.”
This research was published in the international journal ‘Nature Aging’ on May 5, 2025, with Dr. Kwon Yong Tak of KAIST KAIST GSMSE, Ph.D. Candidate Juyeon Kim of KRIBB, and Ph.D. Candidate Myungsun Park of KAIST as co-first authors.
*Paper Title: Quasi-spatial single-cell transcriptome based on physical tissue properties defines early aging associated niche in liver
*DOI: https://doi.org/10.1038/s43587-025-00857-7
This research was supported by several domestic institutions, including the National Research Foundation of Korea, the Korea Health Industry Development Institute (KHIDI), the Korea Research Institute of Bioscience and Biotechnology (KRIBB), KIST, POSCO Science Fellowship, and the Convergence Medical Scientist Training Program.
2025.06.12 View 2359 -
 KAIST sends out Music and Bio-Signs of Professor Kwon Ji-yong, a.k.a. G-Dragon, into Space to Pulsate through Universe and Resonate among Stars
KAIST (President Kwang-Hyung Lee) announced on the 10th of April that it successfully promoted the world’s first ‘Space Sound Source Transmission Project’ based on media art at the KAIST Space Research Institute on April 9th through collaboration between Professor Jinjoon Lee of the Graduate School of Culture Technology, a world-renowned media artist, and the global K-Pop artist, G-Dragon.
This project was proposed as part of the ‘AI Entertech Research Center’ being promoted by KAIST and Galaxy Corporation. It is a project to transmit the message and sound of G-Dragon (real name, Kwon Ji-yong), a singer/song writer affiliated with Galaxy Corporation and a visiting professor in the Department of Mechanical Engineering at KAIST, to space for the first time in the world.
This is a convergence project that combines science, technology, art, and popular music, and is a new form of ‘space culture content’ experiment that connects KAIST’s cutting-edge space technology, Professor Jinjoon Lee’s media art work, and G-Dragon’s voice and sound source containing his latest digital single, "HOME SWEET HOME".
< Photo 1. Professor Jinjoon Lee's Open Your Eyes Project "Iris"'s imagery projected on the 13m space antenna at the Space Research Institute >
This collaboration was planned with the theme of ‘emotional signals that expand the inner universe of humans to the outer universe.’ The image of G-Dragon’s iris was augmented through AI as a window into soul symbolizing his uniqueness and identity, and the new song “Home Sweet Home” was combined as an audio message containing the vibration of that emotion.
This was actually transmitted into space using a next-generation small satellite developed by KAIST Space Research Institute, completing a symbolic performance in which an individual’s inner universe is transmitted to outer space.
Professor Jinjoon Lee’s cinematic media art work “Iris” was unveiled at the site. This work was screened in the world’s first projection mapping method* on KAIST Space Research Institute’s 13m space antenna. This video was created using generative artificial intelligence (AI) technology based on the image of G-Dragon's iris, and combined with sound using the data of the sounds of Emile Bell rings – the bell that holds a thousand years of history, it presented an emotional art experience that transcends time and space.
*Projection Mapping: A technology that projects light and images onto actual structures to create visual changes, and is a method of expression that artistically reinterprets space.
This work is one of the major research achievements of KAIST TX Lab and Professor Lee based on new media technology based on biometric data such as iris, heartbeat, and brain waves.
Professor Jinjoon Lee said, "The iris is a symbol that reflects inner emotions and identity, so much so that it is called the 'mirror of the soul,' and this work sought to express 'the infinite universe seen from the inside of humanity' through G-Dragon's gaze."
< Photo 2. (From left) Professor Jinjoon Lee of the Graduate School of Culture Technology and G-Dragon (Visiting Professor Kwon Ji-yong of the Department of Mechanical Engineering) >
He continued, "The universe is a realm of technology as well as a stage for imagination and emotion, and I look forward to an encounter with the unknown through a new attempt to speak of art in the language of science including AI and imagine science in the form of art." “G-Dragon’s voice and music have now begun their journey to space,” said Yong-ho Choi, Galaxy Corporation’s Chief Happiness Officer (CHO). “This project is an act of leaving music as a legacy for humanity, while also having an important meaning of attempting to communicate with space.” He added, “This is a pioneering step to introduce human culture to space, and it will remain as a monumental performance that opens a new chapter in the history of music comparable to the Beatles.”
Galaxy Corporation is leading the future entertainment technology industry through its collaboration with KAIST, and was recently selected as the only entertainment technology company in a private meeting with Microsoft CEO Nadella. In particular, it is promoting the globalization of AI entertainment technology, receiving praise as a “pioneer of imagination” for new forms of AI entertainment content, including the AI contents for the deceased.
< Photo 3. Photo of G-Dragon's Home Sweet Home being sent into the space via Professor Jinjoon Lee's Space Sound Source Transmission Project >
Through this project, KAIST Space Research Institute presented new possibilities for utilizing satellite technology, and showed a model for science to connect with society in a more popular way.
KAIST President Kwang-Hyung Lee said, “KAIST is a place that always supports new imaginations and challenges,” and added, “We will continue to strive to continue creative research that no one has ever thought of, like this project that combines science, technology, and art.”
In the meantime, Galaxy Corporation, the agency of G-Dragon’s Professor Kwon Ji-yong, is an AI entertainment company that presents a new paradigm based on IP, media, tech, and entertainment convergence technology.
2025.04.10 View 6133
KAIST sends out Music and Bio-Signs of Professor Kwon Ji-yong, a.k.a. G-Dragon, into Space to Pulsate through Universe and Resonate among Stars
KAIST (President Kwang-Hyung Lee) announced on the 10th of April that it successfully promoted the world’s first ‘Space Sound Source Transmission Project’ based on media art at the KAIST Space Research Institute on April 9th through collaboration between Professor Jinjoon Lee of the Graduate School of Culture Technology, a world-renowned media artist, and the global K-Pop artist, G-Dragon.
This project was proposed as part of the ‘AI Entertech Research Center’ being promoted by KAIST and Galaxy Corporation. It is a project to transmit the message and sound of G-Dragon (real name, Kwon Ji-yong), a singer/song writer affiliated with Galaxy Corporation and a visiting professor in the Department of Mechanical Engineering at KAIST, to space for the first time in the world.
This is a convergence project that combines science, technology, art, and popular music, and is a new form of ‘space culture content’ experiment that connects KAIST’s cutting-edge space technology, Professor Jinjoon Lee’s media art work, and G-Dragon’s voice and sound source containing his latest digital single, "HOME SWEET HOME".
< Photo 1. Professor Jinjoon Lee's Open Your Eyes Project "Iris"'s imagery projected on the 13m space antenna at the Space Research Institute >
This collaboration was planned with the theme of ‘emotional signals that expand the inner universe of humans to the outer universe.’ The image of G-Dragon’s iris was augmented through AI as a window into soul symbolizing his uniqueness and identity, and the new song “Home Sweet Home” was combined as an audio message containing the vibration of that emotion.
This was actually transmitted into space using a next-generation small satellite developed by KAIST Space Research Institute, completing a symbolic performance in which an individual’s inner universe is transmitted to outer space.
Professor Jinjoon Lee’s cinematic media art work “Iris” was unveiled at the site. This work was screened in the world’s first projection mapping method* on KAIST Space Research Institute’s 13m space antenna. This video was created using generative artificial intelligence (AI) technology based on the image of G-Dragon's iris, and combined with sound using the data of the sounds of Emile Bell rings – the bell that holds a thousand years of history, it presented an emotional art experience that transcends time and space.
*Projection Mapping: A technology that projects light and images onto actual structures to create visual changes, and is a method of expression that artistically reinterprets space.
This work is one of the major research achievements of KAIST TX Lab and Professor Lee based on new media technology based on biometric data such as iris, heartbeat, and brain waves.
Professor Jinjoon Lee said, "The iris is a symbol that reflects inner emotions and identity, so much so that it is called the 'mirror of the soul,' and this work sought to express 'the infinite universe seen from the inside of humanity' through G-Dragon's gaze."
< Photo 2. (From left) Professor Jinjoon Lee of the Graduate School of Culture Technology and G-Dragon (Visiting Professor Kwon Ji-yong of the Department of Mechanical Engineering) >
He continued, "The universe is a realm of technology as well as a stage for imagination and emotion, and I look forward to an encounter with the unknown through a new attempt to speak of art in the language of science including AI and imagine science in the form of art." “G-Dragon’s voice and music have now begun their journey to space,” said Yong-ho Choi, Galaxy Corporation’s Chief Happiness Officer (CHO). “This project is an act of leaving music as a legacy for humanity, while also having an important meaning of attempting to communicate with space.” He added, “This is a pioneering step to introduce human culture to space, and it will remain as a monumental performance that opens a new chapter in the history of music comparable to the Beatles.”
Galaxy Corporation is leading the future entertainment technology industry through its collaboration with KAIST, and was recently selected as the only entertainment technology company in a private meeting with Microsoft CEO Nadella. In particular, it is promoting the globalization of AI entertainment technology, receiving praise as a “pioneer of imagination” for new forms of AI entertainment content, including the AI contents for the deceased.
< Photo 3. Photo of G-Dragon's Home Sweet Home being sent into the space via Professor Jinjoon Lee's Space Sound Source Transmission Project >
Through this project, KAIST Space Research Institute presented new possibilities for utilizing satellite technology, and showed a model for science to connect with society in a more popular way.
KAIST President Kwang-Hyung Lee said, “KAIST is a place that always supports new imaginations and challenges,” and added, “We will continue to strive to continue creative research that no one has ever thought of, like this project that combines science, technology, and art.”
In the meantime, Galaxy Corporation, the agency of G-Dragon’s Professor Kwon Ji-yong, is an AI entertainment company that presents a new paradigm based on IP, media, tech, and entertainment convergence technology.
2025.04.10 View 6133 -
 KAIST Discovers Molecular Switch that Reverses Cancerous Transformation at the Critical Moment of Transition
< (From left) PhD student Seoyoon D. Jeong, (bottom) Professor Kwang-Hyun Cho, (top) Dr. Dongkwan Shin, Dr. Jeong-Ryeol Gong >
Professor Kwang-Hyun Cho’s research team has recently been highlighted for their work on developing an original technology for cancer reversal treatment that does not kill cancer cells but only changes their characteristics to reverse them to a state similar to normal cells. This time, they have succeeded in revealing for the first time that a molecular switch that can induce cancer reversal at the moment when normal cells change into cancer cells is hidden in the genetic network.
KAIST (President Kwang-Hyung Lee) announced on the 5th of February that Professor Kwang-Hyun Cho's research team of the Department of Bio and Brain Engineering has succeeded in developing a fundamental technology to capture the critical transition phenomenon at the moment when normal cells change into cancer cells and analyze it to discover a molecular switch that can revert cancer cells back into normal cells.
A critical transition is a phenomenon in which a sudden change in state occurs at a specific point in time, like water changing into steam at 100℃. This critical transition phenomenon also occurs in the process in which normal cells change into cancer cells at a specific point in time due to the accumulation of genetic and epigenetic changes.
The research team discovered that normal cells can enter an unstable critical transition state where normal cells and cancer cells coexist just before they change into cancer cells during tumorigenesis, the production or development of tumors, and analyzed this critical transition state using a systems biology method to develop a cancer reversal molecular switch identification technology that can reverse the cancerization process. They then applied this to colon cancer cells and confirmed through molecular cell experiments that cancer cells can recover the characteristics of normal cells.
This is an original technology that automatically infers a computer model of the genetic network that controls the critical transition of cancer development from single-cell RNA sequencing data, and systematically finds molecular switches for cancer reversion by simulation analysis. It is expected that this technology will be applied to the development of reversion therapies for other cancers in the future.
Professor Kwang-Hyun Cho said, "We have discovered a molecular switch that can revert the fate of cancer cells back to a normal state by capturing the moment of critical transition right before normal cells are changed into an irreversible cancerous state."
< Figure 1. Overall conceptual framework of the technology that automatically constructs a molecular regulatory network from single-cell RNA sequencing data of colon cancer cells to discover molecular switches for cancer reversion through computer simulation analysis. Professor Kwang-Hyun Cho's research team established a fundamental technology for automatic construction of a computer model of a core gene network by analyzing the entire process of tumorigenesis of colon cells turning into cancer cells, and developed an original technology for discovering the molecular switches that can induce cancer cell reversal through attractor landscape analysis. >
He continued, "In particular, this study has revealed in detail, at the genetic network level, what changes occur within cells behind the process of cancer development, which has been considered a mystery until now." He emphasized, "This is the first study to reveal that an important clue that can revert the fate of tumorigenesis is hidden at this very critical moment of change."
< Figure 2. Identification of tumor transition state using single-cell RNA sequencing data from colorectal cancer. Using single-cell RNA sequencing data from colorectal cancer patient-derived organoids for normal and cancerous tissues, a critical transition was identified in which normal and cancerous cells coexist and instability increases (a-d). The critical transition was confirmed to show intermediate levels of major phenotypic features related to cancer or normal tissues that are indicative of the states between the normal and cancerous cells (e). >
The results of this study, conducted by KAIST Dr. Dongkwan Shin (currently at the National Cancer Center), Dr. Jeong-Ryeol Gong, and doctoral student Seoyoon D. Jeong jointly with a research team at Seoul National University that provided the organoids (in vitro cultured tissues) from colon cancer patient, were published as an online paper in the international journal ‘Advanced Science’ published by Wiley on January 22nd.
(Paper title: Attractor landscape analysis reveals a reversion switch in the transition of colorectal tumorigenesis) (DOI: https://doi.org/10.1002/advs.202412503)
< Figure 3. Reconstruction of a dynamic network model for the transition state of colorectal cancer.
A new technology was established to build a gene network computer model that can simulate the dynamic changes between genes by integrating single-cell RNA sequencing data and existing experimental results on gene-to-gene interactions in the critical transition of cancer. (a). Using this technology, a gene network computer model for the critical transition of colorectal cancer was constructed, and the distribution of attractors representing normal and cancer cell phenotypes was investigated through attractor landscape analysis (b-e). >
This study was conducted with the support of the National Research Foundation of Korea under the Ministry of Science and ICT through the Mid-Career Researcher Program and Basic Research Laboratory Program and the Disease-Centered Translational Research Project of the Korea Health Industry Development Institute (KHIDI) of the Ministry of Health and Welfare.
< Figure 4. Quantification of attractor landscapes and discovery of transcription factors for cancer reversibility through perturbation simulation analysis. A methodology for implementing discontinuous attractor landscapes continuously from a computer model of gene networks and quantifying them as cancer scores was introduced (a), and attractor landscapes for the critical transition of colorectal cancer were secured (b-d). By tracking the change patterns of normal and cancer cell attractors through perturbation simulation analysis for each gene, the optimal combination of transcription factors for cancer reversion was discovered (e-h). This was confirmed in various parameter combinations as well (i). >
< Figure 5. Identification and experimental validation of the optimal target gene for cancer reversion. Among the common target genes of the discovered transcription factor combinations, we identified cancer reversing molecular switches that are predicted to suppress cancer cell proliferation and restore the characteristics of normal colon cells (a-d). When inhibitors for the molecular switches were treated to organoids derived from colon cancer patients, it was confirmed that cancer cell proliferation was suppressed and the expression of key genes related to cancer development was inhibited (e-h), and a group of genes related to normal colon epithelium was activated and transformed into a state similar to normal colon cells (i-j). >
< Figure 6. Schematic diagram of the research results. Professor Kwang-Hyun Cho's research team developed an original technology to systematically discover key molecular switches that can induce reversion of colon cancer cells through a systems biology approach using an attractor landscape analysis of a genetic network model for the critical transition at the moment of transformation from normal cells to cancer cells, and verified the reversing effect of actual colon cancer through cellular experiments. >
2025.02.05 View 29895
KAIST Discovers Molecular Switch that Reverses Cancerous Transformation at the Critical Moment of Transition
< (From left) PhD student Seoyoon D. Jeong, (bottom) Professor Kwang-Hyun Cho, (top) Dr. Dongkwan Shin, Dr. Jeong-Ryeol Gong >
Professor Kwang-Hyun Cho’s research team has recently been highlighted for their work on developing an original technology for cancer reversal treatment that does not kill cancer cells but only changes their characteristics to reverse them to a state similar to normal cells. This time, they have succeeded in revealing for the first time that a molecular switch that can induce cancer reversal at the moment when normal cells change into cancer cells is hidden in the genetic network.
KAIST (President Kwang-Hyung Lee) announced on the 5th of February that Professor Kwang-Hyun Cho's research team of the Department of Bio and Brain Engineering has succeeded in developing a fundamental technology to capture the critical transition phenomenon at the moment when normal cells change into cancer cells and analyze it to discover a molecular switch that can revert cancer cells back into normal cells.
A critical transition is a phenomenon in which a sudden change in state occurs at a specific point in time, like water changing into steam at 100℃. This critical transition phenomenon also occurs in the process in which normal cells change into cancer cells at a specific point in time due to the accumulation of genetic and epigenetic changes.
The research team discovered that normal cells can enter an unstable critical transition state where normal cells and cancer cells coexist just before they change into cancer cells during tumorigenesis, the production or development of tumors, and analyzed this critical transition state using a systems biology method to develop a cancer reversal molecular switch identification technology that can reverse the cancerization process. They then applied this to colon cancer cells and confirmed through molecular cell experiments that cancer cells can recover the characteristics of normal cells.
This is an original technology that automatically infers a computer model of the genetic network that controls the critical transition of cancer development from single-cell RNA sequencing data, and systematically finds molecular switches for cancer reversion by simulation analysis. It is expected that this technology will be applied to the development of reversion therapies for other cancers in the future.
Professor Kwang-Hyun Cho said, "We have discovered a molecular switch that can revert the fate of cancer cells back to a normal state by capturing the moment of critical transition right before normal cells are changed into an irreversible cancerous state."
< Figure 1. Overall conceptual framework of the technology that automatically constructs a molecular regulatory network from single-cell RNA sequencing data of colon cancer cells to discover molecular switches for cancer reversion through computer simulation analysis. Professor Kwang-Hyun Cho's research team established a fundamental technology for automatic construction of a computer model of a core gene network by analyzing the entire process of tumorigenesis of colon cells turning into cancer cells, and developed an original technology for discovering the molecular switches that can induce cancer cell reversal through attractor landscape analysis. >
He continued, "In particular, this study has revealed in detail, at the genetic network level, what changes occur within cells behind the process of cancer development, which has been considered a mystery until now." He emphasized, "This is the first study to reveal that an important clue that can revert the fate of tumorigenesis is hidden at this very critical moment of change."
< Figure 2. Identification of tumor transition state using single-cell RNA sequencing data from colorectal cancer. Using single-cell RNA sequencing data from colorectal cancer patient-derived organoids for normal and cancerous tissues, a critical transition was identified in which normal and cancerous cells coexist and instability increases (a-d). The critical transition was confirmed to show intermediate levels of major phenotypic features related to cancer or normal tissues that are indicative of the states between the normal and cancerous cells (e). >
The results of this study, conducted by KAIST Dr. Dongkwan Shin (currently at the National Cancer Center), Dr. Jeong-Ryeol Gong, and doctoral student Seoyoon D. Jeong jointly with a research team at Seoul National University that provided the organoids (in vitro cultured tissues) from colon cancer patient, were published as an online paper in the international journal ‘Advanced Science’ published by Wiley on January 22nd.
(Paper title: Attractor landscape analysis reveals a reversion switch in the transition of colorectal tumorigenesis) (DOI: https://doi.org/10.1002/advs.202412503)
< Figure 3. Reconstruction of a dynamic network model for the transition state of colorectal cancer.
A new technology was established to build a gene network computer model that can simulate the dynamic changes between genes by integrating single-cell RNA sequencing data and existing experimental results on gene-to-gene interactions in the critical transition of cancer. (a). Using this technology, a gene network computer model for the critical transition of colorectal cancer was constructed, and the distribution of attractors representing normal and cancer cell phenotypes was investigated through attractor landscape analysis (b-e). >
This study was conducted with the support of the National Research Foundation of Korea under the Ministry of Science and ICT through the Mid-Career Researcher Program and Basic Research Laboratory Program and the Disease-Centered Translational Research Project of the Korea Health Industry Development Institute (KHIDI) of the Ministry of Health and Welfare.
< Figure 4. Quantification of attractor landscapes and discovery of transcription factors for cancer reversibility through perturbation simulation analysis. A methodology for implementing discontinuous attractor landscapes continuously from a computer model of gene networks and quantifying them as cancer scores was introduced (a), and attractor landscapes for the critical transition of colorectal cancer were secured (b-d). By tracking the change patterns of normal and cancer cell attractors through perturbation simulation analysis for each gene, the optimal combination of transcription factors for cancer reversion was discovered (e-h). This was confirmed in various parameter combinations as well (i). >
< Figure 5. Identification and experimental validation of the optimal target gene for cancer reversion. Among the common target genes of the discovered transcription factor combinations, we identified cancer reversing molecular switches that are predicted to suppress cancer cell proliferation and restore the characteristics of normal colon cells (a-d). When inhibitors for the molecular switches were treated to organoids derived from colon cancer patients, it was confirmed that cancer cell proliferation was suppressed and the expression of key genes related to cancer development was inhibited (e-h), and a group of genes related to normal colon epithelium was activated and transformed into a state similar to normal colon cells (i-j). >
< Figure 6. Schematic diagram of the research results. Professor Kwang-Hyun Cho's research team developed an original technology to systematically discover key molecular switches that can induce reversion of colon cancer cells through a systems biology approach using an attractor landscape analysis of a genetic network model for the critical transition at the moment of transformation from normal cells to cancer cells, and verified the reversing effect of actual colon cancer through cellular experiments. >
2025.02.05 View 29895 -
 KAIST to Collaborate with AT&C to Take Dominance over Dementia
< Photo 1. (From left) KAIST Dean of the College of Natural Sciences Daesoo Kim, KAIST President Kwang Hyung Lee, AT&C Chairman Ki Tae Lee, AT&C CEO Jong-won Lee >
KAIST (President Kwang Hyung Lee) announced on January 9th that it signed a memorandum of understanding for a comprehensive mutual cooperation with AT&C (CEO Jong-won Lee) at its Seoul Dogok Campus to expand research investment and industry-academia cooperation in preparation for the future cutting-edge digital bio era.
Senile dementia is a rapidly increasing brain disease that affects 10% of the elderly population aged 65 and older, and approximately 38% of those aged 85 and older suffer from dementia. Alzheimer's disease is the most common dementia in the elderly and its prevalence has been increasing rapidly in the population of over 40 years of age. However, an effective treatment is yet to be found.
The Korean government is investing a total of KRW 1.1 trillion in dementia R&D projects from 2020 to 2029, with the goal of reducing the rate of increase of dementia patients by 50%. Since it takes a lot of time and money to develop effective and affordable medicinal dementia treatments, it is urgent to work on the development of digital treatments for dementia that can be applied more quickly.
AT&C, a digital healthcare company, has already received approval from the Ministry of Food and Drug Safety (MFDS) for its device for antidepressant treatment based on transcranial magnetic stimulation (TMS) using magnetic fields and is selling it domestically and internationally. In addition, it has developed the first Alzheimer's dementia treatment device in Korea and received MFDS approval for clinical trials. After passing phase 1 to evaluate safety and phase 2 to test efficacy on some patients, it is currently conducting phase 3 clinical trials to test efficacy on a larger group of patients.
This dementia treatment device is equipped with a system that combines non-invasive electronic stimulations (TMS electromagnetic stimulator) and digital therapeutic prescription (cognitive learning programs) to provide precise, automated treatment by applying AI image analysis and robotics technology.
Through this agreement, KAIST and AT&C have agreed to cooperate with each other in the development of innovative digital treatment equipment for brain diseases. Through research collaboration with KAIST, AT&C will be able to develop technology that can be widely applied to Parkinson's disease, stroke, mild cognitive impairment, sleep disorders, etc., and will develop portable equipment that can improve brain function and prevent dementia at home by utilizing KAIST's wearable technology.
To this end, AT&C plans to establish a digital healthcare research center at KAIST by supporting research personnel and research expenses worth approximately 3 billion won with the goal of developing cutting-edge digital equipment within 3 years.
The digital equipment market is expected to grow at a compounded annual growth rate of 22.1% from 2023 to 2033, reaching a market size of $1.9209 trillion by 2033.
< Photo 2. (From left) Dean of the KAIST College of Natural Sciences Daesoo Kim, Professor Young-joon Lee, Professor Minee Choi of the KAIST Department of Brain and Cognitive Sciences, KAIST President Kwang Hyung Lee, Chairman Ki Tae Lee, CEO Jong-won Lee, and Headquarters Director Ki-yong Na of AT&C >
CEO Jong-won Lee said, “AT&C is playing a leading role in the treatment of Alzheimer’s disease using TMS (transcranial magnetic stimulation) technology. Through this agreement with KAIST, we will do our best to create a new paradigm for brain disease treatment and become a platform company that can lead future medical devices and medical technology.”
Former Samsung Electronics Vice Chairman Ki Tae Lee, a strong supporter of this R&D project, said, “Through this agreement with KAIST, we plan to prepare for a new future by combining the technologies AT&C has developed so far with KAIST’s innovative and differentiated technologies.”
KAIST President Kwang Hyung Lee emphasized, “Through this collaboration, KAIST expects to build a world-class digital therapeutics infrastructure for treating brain diseases and contribute greatly to further strengthening Korea’s competitiveness in the biomedical field.”
The signing ceremony was attended by KAIST President Kwang Hyung Lee, the Dean of KAIST College of Natural Sciences Daesoo Kim, AT&C CEO Lee Jong-won, and the current Chairman of AT&C, Ki Tae Lee, former Vice Chairman of Samsung Electronics.
2025.01.09 View 6386
KAIST to Collaborate with AT&C to Take Dominance over Dementia
< Photo 1. (From left) KAIST Dean of the College of Natural Sciences Daesoo Kim, KAIST President Kwang Hyung Lee, AT&C Chairman Ki Tae Lee, AT&C CEO Jong-won Lee >
KAIST (President Kwang Hyung Lee) announced on January 9th that it signed a memorandum of understanding for a comprehensive mutual cooperation with AT&C (CEO Jong-won Lee) at its Seoul Dogok Campus to expand research investment and industry-academia cooperation in preparation for the future cutting-edge digital bio era.
Senile dementia is a rapidly increasing brain disease that affects 10% of the elderly population aged 65 and older, and approximately 38% of those aged 85 and older suffer from dementia. Alzheimer's disease is the most common dementia in the elderly and its prevalence has been increasing rapidly in the population of over 40 years of age. However, an effective treatment is yet to be found.
The Korean government is investing a total of KRW 1.1 trillion in dementia R&D projects from 2020 to 2029, with the goal of reducing the rate of increase of dementia patients by 50%. Since it takes a lot of time and money to develop effective and affordable medicinal dementia treatments, it is urgent to work on the development of digital treatments for dementia that can be applied more quickly.
AT&C, a digital healthcare company, has already received approval from the Ministry of Food and Drug Safety (MFDS) for its device for antidepressant treatment based on transcranial magnetic stimulation (TMS) using magnetic fields and is selling it domestically and internationally. In addition, it has developed the first Alzheimer's dementia treatment device in Korea and received MFDS approval for clinical trials. After passing phase 1 to evaluate safety and phase 2 to test efficacy on some patients, it is currently conducting phase 3 clinical trials to test efficacy on a larger group of patients.
This dementia treatment device is equipped with a system that combines non-invasive electronic stimulations (TMS electromagnetic stimulator) and digital therapeutic prescription (cognitive learning programs) to provide precise, automated treatment by applying AI image analysis and robotics technology.
Through this agreement, KAIST and AT&C have agreed to cooperate with each other in the development of innovative digital treatment equipment for brain diseases. Through research collaboration with KAIST, AT&C will be able to develop technology that can be widely applied to Parkinson's disease, stroke, mild cognitive impairment, sleep disorders, etc., and will develop portable equipment that can improve brain function and prevent dementia at home by utilizing KAIST's wearable technology.
To this end, AT&C plans to establish a digital healthcare research center at KAIST by supporting research personnel and research expenses worth approximately 3 billion won with the goal of developing cutting-edge digital equipment within 3 years.
The digital equipment market is expected to grow at a compounded annual growth rate of 22.1% from 2023 to 2033, reaching a market size of $1.9209 trillion by 2033.
< Photo 2. (From left) Dean of the KAIST College of Natural Sciences Daesoo Kim, Professor Young-joon Lee, Professor Minee Choi of the KAIST Department of Brain and Cognitive Sciences, KAIST President Kwang Hyung Lee, Chairman Ki Tae Lee, CEO Jong-won Lee, and Headquarters Director Ki-yong Na of AT&C >
CEO Jong-won Lee said, “AT&C is playing a leading role in the treatment of Alzheimer’s disease using TMS (transcranial magnetic stimulation) technology. Through this agreement with KAIST, we will do our best to create a new paradigm for brain disease treatment and become a platform company that can lead future medical devices and medical technology.”
Former Samsung Electronics Vice Chairman Ki Tae Lee, a strong supporter of this R&D project, said, “Through this agreement with KAIST, we plan to prepare for a new future by combining the technologies AT&C has developed so far with KAIST’s innovative and differentiated technologies.”
KAIST President Kwang Hyung Lee emphasized, “Through this collaboration, KAIST expects to build a world-class digital therapeutics infrastructure for treating brain diseases and contribute greatly to further strengthening Korea’s competitiveness in the biomedical field.”
The signing ceremony was attended by KAIST President Kwang Hyung Lee, the Dean of KAIST College of Natural Sciences Daesoo Kim, AT&C CEO Lee Jong-won, and the current Chairman of AT&C, Ki Tae Lee, former Vice Chairman of Samsung Electronics.
2025.01.09 View 6386 -
 KAIST Opens Newly Expanded Center for Contemplative Research in Collaboration with Brain and Cognitive Sciences Department
KAIST (represented by President Kwang Hyung Lee) announced on January 2nd that it would hold an opening ceremony for the expanded KAIST Center for Contemplative Research (Director Wan Doo Kim) at the Creativity Learning Building on its Daejeon campus on January 3 (Friday).
Established in 2018 with the mission of "integrating meditation and science for the happiness and prosperity of humanity," the KAIST Center for Contemplative Research has been expanding its scope of research into the neuroscience of meditation and training empathetic educators who will lead the field of meditation science in collaboration with the Brain and Cognitive Sciences Department, which was established in 2022.
Supported by the Plato Academy Foundation and with funding from SK Discovery for the facility’s expansion, the center now occupies an extended space on the 5th floor of the Creativity Learning Center. The new facilities include: ▲ Advanced Research Equipment ▲ Meditation Science Laboratories ▲ VR/XR-Based Meditation Experience Rooms ▲ A Large Digital Art Meditation Hall ▲ Personal Meditation Halls.
Particularly, the center plans to conduct next-generation meditation research using cutting-edge technologies such as: ▲ Brain-Computer Interface Technology ▲ Meditation Wearable Devices ▲ Metaverse-Based Meditation Environments.
The opening ceremony, scheduled for the morning of January 3 (Friday), was attended by key figures, including Plato Academy Foundation Chairman Chang-Won Choi, MindLab CEO Professor Seong-Taek Cho, Bosung Group Vice President Byung-Chul Lee, and KAIST President Kwang Hyung Lee.
The event began with a national moment of silence to honor the victims of the recent Jeju Air passenger accident. It included a progress report by the center director, a lecture by Professor Jaeseung Jeong, panel discussions, and more.
Following a tour of the expanded facilities, the center hosted a 20-minute hands-on meditation science session using *Looxid Labs EEG devices for the first 50 participants.
*Looxid Labs EEG Device: A real-time brainwave measurement device developed by KAIST startup Looxid Labs that enables users to experience efficient and AI-powered data-driven meditation science practice (Looxid Labs website: https://looxidlabs.com/).
During the ceremony, Director of the Center for Contemplative Research Wan Doo Kim presented on "The Mission, Vision, and Future of the KAIST Center for Contemplative Research." Yujin Lee, a combined master’s and doctoral researcher from the Brain and Cognitive Sciences Department, shared insights on "The Latest Trends in Meditation Science Research."
A panel discussion and Q&A session on "The Convergence of Meditation and Brain and Cognitive Sciences" followed featuring Professors Jaeseung Jeong, HyungDong Park (Brain and Cognitive Sciences), and Jiyoung Park (Digital Humanities and Social Sciences).
Director Wan Doo Kim commented, “With this expanded opening, we aim to offer advanced meditation programs integrating brain and cognitive sciences and cutting-edge technology not only to KAIST members but also to the general public interested in meditation. We will continue to dedicate ourselves to interdisciplinary research between meditation and science.”
2025.01.03 View 5318
KAIST Opens Newly Expanded Center for Contemplative Research in Collaboration with Brain and Cognitive Sciences Department
KAIST (represented by President Kwang Hyung Lee) announced on January 2nd that it would hold an opening ceremony for the expanded KAIST Center for Contemplative Research (Director Wan Doo Kim) at the Creativity Learning Building on its Daejeon campus on January 3 (Friday).
Established in 2018 with the mission of "integrating meditation and science for the happiness and prosperity of humanity," the KAIST Center for Contemplative Research has been expanding its scope of research into the neuroscience of meditation and training empathetic educators who will lead the field of meditation science in collaboration with the Brain and Cognitive Sciences Department, which was established in 2022.
Supported by the Plato Academy Foundation and with funding from SK Discovery for the facility’s expansion, the center now occupies an extended space on the 5th floor of the Creativity Learning Center. The new facilities include: ▲ Advanced Research Equipment ▲ Meditation Science Laboratories ▲ VR/XR-Based Meditation Experience Rooms ▲ A Large Digital Art Meditation Hall ▲ Personal Meditation Halls.
Particularly, the center plans to conduct next-generation meditation research using cutting-edge technologies such as: ▲ Brain-Computer Interface Technology ▲ Meditation Wearable Devices ▲ Metaverse-Based Meditation Environments.
The opening ceremony, scheduled for the morning of January 3 (Friday), was attended by key figures, including Plato Academy Foundation Chairman Chang-Won Choi, MindLab CEO Professor Seong-Taek Cho, Bosung Group Vice President Byung-Chul Lee, and KAIST President Kwang Hyung Lee.
The event began with a national moment of silence to honor the victims of the recent Jeju Air passenger accident. It included a progress report by the center director, a lecture by Professor Jaeseung Jeong, panel discussions, and more.
Following a tour of the expanded facilities, the center hosted a 20-minute hands-on meditation science session using *Looxid Labs EEG devices for the first 50 participants.
*Looxid Labs EEG Device: A real-time brainwave measurement device developed by KAIST startup Looxid Labs that enables users to experience efficient and AI-powered data-driven meditation science practice (Looxid Labs website: https://looxidlabs.com/).
During the ceremony, Director of the Center for Contemplative Research Wan Doo Kim presented on "The Mission, Vision, and Future of the KAIST Center for Contemplative Research." Yujin Lee, a combined master’s and doctoral researcher from the Brain and Cognitive Sciences Department, shared insights on "The Latest Trends in Meditation Science Research."
A panel discussion and Q&A session on "The Convergence of Meditation and Brain and Cognitive Sciences" followed featuring Professors Jaeseung Jeong, HyungDong Park (Brain and Cognitive Sciences), and Jiyoung Park (Digital Humanities and Social Sciences).
Director Wan Doo Kim commented, “With this expanded opening, we aim to offer advanced meditation programs integrating brain and cognitive sciences and cutting-edge technology not only to KAIST members but also to the general public interested in meditation. We will continue to dedicate ourselves to interdisciplinary research between meditation and science.”
2025.01.03 View 5318 -
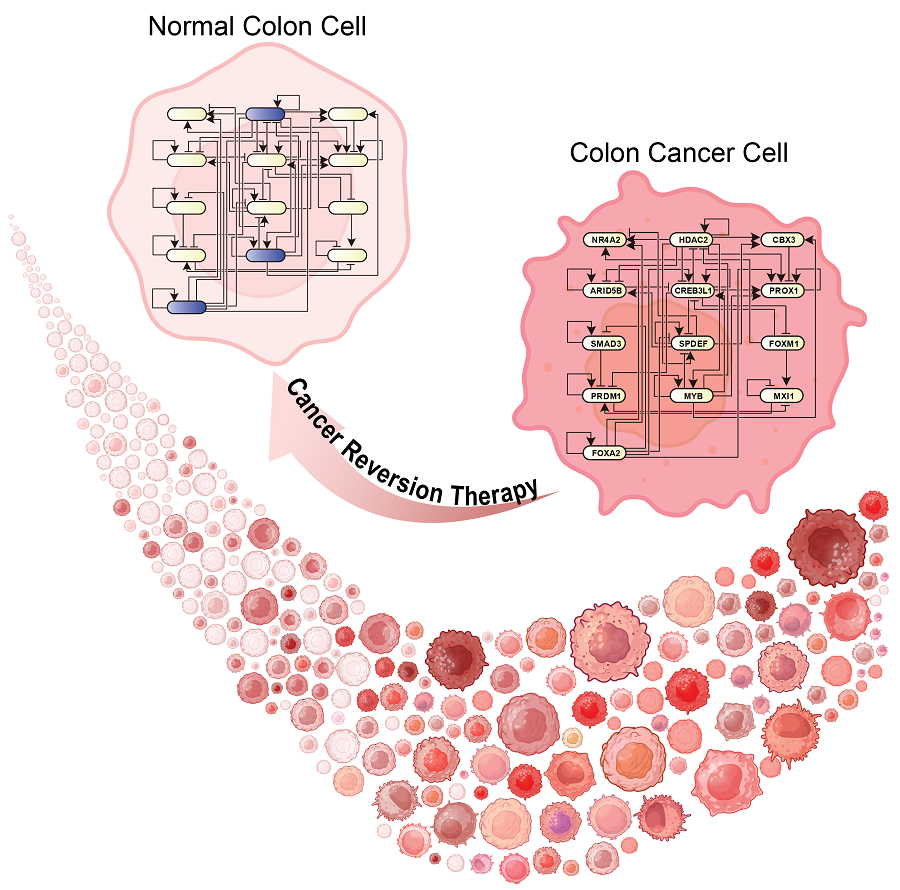 KAIST Develops Foundational Technology to Revert Cancer Cells to Normal Cells
Despite the development of numerous cancer treatment technologies, the common goal of current cancer therapies is to eliminate cancer cells. This approach, however, faces fundamental limitations, including cancer cells developing resistance and returning, as well as severe side effects from the destruction of healthy cells.
< (From top left) Bio and Brain Engineering PhD candidates Juhee Kim, Jeong-Ryeol Gong, Chun-Kyung Lee, and Hoon-Min Kim posed for a group photo with Professor Kwang-Hyun Cho >
KAIST (represented by President Kwang Hyung Lee) announced on the 20th of December that a research team led by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering has developed a groundbreaking technology that can treat colon cancer by converting cancer cells into a state resembling normal colon cells without killing them, thus avoiding side effects.
The research team focused on the observation that during the oncogenesis process, normal cells regress along their differentiation trajectory. Building on this insight, they developed a technology to create a digital twin of the gene network associated with the differentiation trajectory of normal cells.
< Figure 1. Technology for creating a digital twin of a gene network from single-cell transcriptome data of a normal cell differentiation trajectory. Professor Kwang-Hyun Cho's research team developed a digital twin creation technology that precisely observes the dynamics of gene regulatory relationships during the process of normal cells differentiating along a differentiation trajectory and analyzes the relationships among key genes to build a mathematical model that can be simulated (A-F). In addition, they developed a technology to discover key regulatory factors that control the differentiation trajectory of normal cells by simulating and analyzing this digital twin. >
< Figure 2. Digital twin simulation simulating the differentiation trajectory of normal colon cells. The dynamics of single-cell transcriptome data for the differentiation trajectory of normal colon cells were analyzed (A) and a digital twin of the gene network was developed representing the regulatory relationships of key genes in this differentiation trajectory (B). The simulation results of the digital twin confirm that it readily reproduces the dynamics of single-cell transcriptome data (C, D). >
Through simulation analysis, the team systematically identified master molecular switches that induce normal cell differentiation. When these switches were applied to colon cancer cells, the cancer cells reverted to a normal-like state, a result confirmed through molecular and cellular experiments as well as animal studies.
< Figure 3. Discovery of top-level key control factors that induce differentiation of normal colon cells. By applying control factor discovery technology to the digital twin model, three genes, HDAC2, FOXA2, and MYB, were discovered as key control factors that induce differentiation of normal colon cells (A, B). The results of simulation analysis of the regulatory effects of the discovered control factors through the digital twin confirmed that they could induce complete differentiation of colon cells (C). >
< Figure 4. Verification of the effect of the key control factors discovered using colon cancer cells and animal experiments on the reversibility of colon cancer. The key control factors of the normal colon cell differentiation trajectory discovered through digital twin simulation analysis were applied to actual colon cancer cells and colon cancer mouse animal models to experimentally verify the effect of cancer reversibility. The key control factors significantly reduced the proliferation of three colon cancer cell lines (A), and this was confirmed in the same way in animal models (B-D). >
This research demonstrates that cancer cell reversion can be systematically achieved by analyzing and utilizing the digital twin of the cancer cell gene network, rather than relying on serendipitous discoveries. The findings hold significant promise for developing reversible cancer therapies that can be applied to various types of cancer.
< Figure 5. The change in overall gene expression was confirmed through the regulation of the identified key regulatory factors, which converted the state of colon cancer cells to that of normal colon cells. The transcriptomes of colon cancer tissues and normal colon tissues from more than 400 colon cancer patients were compared with the transcriptomes of colon cancer cell lines and reversible colon cancer cell lines, respectively. The comparison results confirmed that the regulation of the identified key regulatory factors converted all three colon cancer cell lines to a state similar to the transcriptome expression of normal colon tissues. >
Professor Kwang-Hyun Cho remarked, "The fact that cancer cells can be converted back to normal cells is an astonishing phenomenon. This study proves that such reversion can be systematically induced."
He further emphasized, "This research introduces the novel concept of reversible cancer therapy by reverting cancer cells to normal cells. It also develops foundational technology for identifying targets for cancer reversion through the systematic analysis of normal cell differentiation trajectories."
This research included contributions from Jeong-Ryeol Gong, Chun-Kyung Lee, Hoon-Min Kim, Juhee Kim, and Jaeog Jeon, and was published in the online edition of the international journal Advanced Science by Wiley on December 11. (Title: “Control of Cellular Differentiation Trajectories for Cancer Reversion”) DOI: https://doi.org/10.1002/advs.202402132
< Figure 6. Schematic diagram of the research results. Professor Kwang-Hyun Cho's research team developed a source technology to systematically discover key control factors that can induce reversibility of colon cancer cells through a systems biology approach and a digital twin simulation analysis of the differentiation trajectory of normal colon cells, and verified the effects of reversion on actual colon cancer through molecular cell experiments and animal experiments. >
The study was supported by the Ministry of Science and ICT and the National Research Foundation of Korea through the Mid-Career Researcher Program and Basic Research Laboratory Program. The research findings have been transferred to BioRevert Inc., where they will be used for the development of practical cancer reversion therapies.
2024.12.23 View 107155
KAIST Develops Foundational Technology to Revert Cancer Cells to Normal Cells
Despite the development of numerous cancer treatment technologies, the common goal of current cancer therapies is to eliminate cancer cells. This approach, however, faces fundamental limitations, including cancer cells developing resistance and returning, as well as severe side effects from the destruction of healthy cells.
< (From top left) Bio and Brain Engineering PhD candidates Juhee Kim, Jeong-Ryeol Gong, Chun-Kyung Lee, and Hoon-Min Kim posed for a group photo with Professor Kwang-Hyun Cho >
KAIST (represented by President Kwang Hyung Lee) announced on the 20th of December that a research team led by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering has developed a groundbreaking technology that can treat colon cancer by converting cancer cells into a state resembling normal colon cells without killing them, thus avoiding side effects.
The research team focused on the observation that during the oncogenesis process, normal cells regress along their differentiation trajectory. Building on this insight, they developed a technology to create a digital twin of the gene network associated with the differentiation trajectory of normal cells.
< Figure 1. Technology for creating a digital twin of a gene network from single-cell transcriptome data of a normal cell differentiation trajectory. Professor Kwang-Hyun Cho's research team developed a digital twin creation technology that precisely observes the dynamics of gene regulatory relationships during the process of normal cells differentiating along a differentiation trajectory and analyzes the relationships among key genes to build a mathematical model that can be simulated (A-F). In addition, they developed a technology to discover key regulatory factors that control the differentiation trajectory of normal cells by simulating and analyzing this digital twin. >
< Figure 2. Digital twin simulation simulating the differentiation trajectory of normal colon cells. The dynamics of single-cell transcriptome data for the differentiation trajectory of normal colon cells were analyzed (A) and a digital twin of the gene network was developed representing the regulatory relationships of key genes in this differentiation trajectory (B). The simulation results of the digital twin confirm that it readily reproduces the dynamics of single-cell transcriptome data (C, D). >
Through simulation analysis, the team systematically identified master molecular switches that induce normal cell differentiation. When these switches were applied to colon cancer cells, the cancer cells reverted to a normal-like state, a result confirmed through molecular and cellular experiments as well as animal studies.
< Figure 3. Discovery of top-level key control factors that induce differentiation of normal colon cells. By applying control factor discovery technology to the digital twin model, three genes, HDAC2, FOXA2, and MYB, were discovered as key control factors that induce differentiation of normal colon cells (A, B). The results of simulation analysis of the regulatory effects of the discovered control factors through the digital twin confirmed that they could induce complete differentiation of colon cells (C). >
< Figure 4. Verification of the effect of the key control factors discovered using colon cancer cells and animal experiments on the reversibility of colon cancer. The key control factors of the normal colon cell differentiation trajectory discovered through digital twin simulation analysis were applied to actual colon cancer cells and colon cancer mouse animal models to experimentally verify the effect of cancer reversibility. The key control factors significantly reduced the proliferation of three colon cancer cell lines (A), and this was confirmed in the same way in animal models (B-D). >
This research demonstrates that cancer cell reversion can be systematically achieved by analyzing and utilizing the digital twin of the cancer cell gene network, rather than relying on serendipitous discoveries. The findings hold significant promise for developing reversible cancer therapies that can be applied to various types of cancer.
< Figure 5. The change in overall gene expression was confirmed through the regulation of the identified key regulatory factors, which converted the state of colon cancer cells to that of normal colon cells. The transcriptomes of colon cancer tissues and normal colon tissues from more than 400 colon cancer patients were compared with the transcriptomes of colon cancer cell lines and reversible colon cancer cell lines, respectively. The comparison results confirmed that the regulation of the identified key regulatory factors converted all three colon cancer cell lines to a state similar to the transcriptome expression of normal colon tissues. >
Professor Kwang-Hyun Cho remarked, "The fact that cancer cells can be converted back to normal cells is an astonishing phenomenon. This study proves that such reversion can be systematically induced."
He further emphasized, "This research introduces the novel concept of reversible cancer therapy by reverting cancer cells to normal cells. It also develops foundational technology for identifying targets for cancer reversion through the systematic analysis of normal cell differentiation trajectories."
This research included contributions from Jeong-Ryeol Gong, Chun-Kyung Lee, Hoon-Min Kim, Juhee Kim, and Jaeog Jeon, and was published in the online edition of the international journal Advanced Science by Wiley on December 11. (Title: “Control of Cellular Differentiation Trajectories for Cancer Reversion”) DOI: https://doi.org/10.1002/advs.202402132
< Figure 6. Schematic diagram of the research results. Professor Kwang-Hyun Cho's research team developed a source technology to systematically discover key control factors that can induce reversibility of colon cancer cells through a systems biology approach and a digital twin simulation analysis of the differentiation trajectory of normal colon cells, and verified the effects of reversion on actual colon cancer through molecular cell experiments and animal experiments. >
The study was supported by the Ministry of Science and ICT and the National Research Foundation of Korea through the Mid-Career Researcher Program and Basic Research Laboratory Program. The research findings have been transferred to BioRevert Inc., where they will be used for the development of practical cancer reversion therapies.
2024.12.23 View 107155 -
 KAIST Proposes a New Way to Circumvent a Long-time Frustration in Neural Computing
The human brain begins learning through spontaneous random activities even before it receives sensory information from the external world. The technology developed by the KAIST research team enables much faster and more accurate learning when exposed to actual data by pre-learning random information in a brain-mimicking artificial neural network, and is expected to be a breakthrough in the development of brain-based artificial intelligence and neuromorphic computing technology in the future.
KAIST (President Kwang-Hyung Lee) announced on the 16th of December that Professor Se-Bum Paik 's research team in the Department of Brain Cognitive Sciences solved the weight transport problem*, a long-standing challenge in neural network learning, and through this, explained the principles that enable resource-efficient learning in biological brain neural networks.
*Weight transport problem: This is the biggest obstacle to the development of artificial intelligence that mimics the biological brain. It is the fundamental reason why large-scale memory and computational work are required in the learning of general artificial neural networks, unlike biological brains.
Over the past several decades, the development of artificial intelligence has been based on error backpropagation learning proposed by Geoffery Hinton, who won the Nobel Prize in Physics this year. However, error backpropagation learning was thought to be impossible in biological brains because it requires the unrealistic assumption that individual neurons must know all the connected information across multiple layers in order to calculate the error signal for learning.
< Figure 1. Illustration depicting the method of random noise training and its effects >
This difficult problem, called the weight transport problem, was raised by Francis Crick, who won the Nobel Prize in Physiology or Medicine for the discovery of the structure of DNA, after the error backpropagation learning was proposed by Hinton in 1986. Since then, it has been considered the reason why the operating principles of natural neural networks and artificial neural networks will forever be fundamentally different.
At the borderline of artificial intelligence and neuroscience, researchers including Hinton have continued to attempt to create biologically plausible models that can implement the learning principles of the brain by solving the weight transport problem.
In 2016, a joint research team from Oxford University and DeepMind in the UK first proposed the concept of error backpropagation learning being possible without weight transport, drawing attention from the academic world. However, biologically plausible error backpropagation learning without weight transport was inefficient, with slow learning speeds and low accuracy, making it difficult to apply in reality.
KAIST research team noted that the biological brain begins learning through internal spontaneous random neural activity even before experiencing external sensory experiences. To mimic this, the research team pre-trained a biologically plausible neural network without weight transport with meaningless random information (random noise).
As a result, they showed that the symmetry of the forward and backward neural cell connections of the neural network, which is an essential condition for error backpropagation learning, can be created. In other words, learning without weight transport is possible through random pre-training.
< Figure 2. Illustration depicting the meta-learning effect of random noise training >
The research team revealed that learning random information before learning actual data has the property of meta-learning, which is ‘learning how to learn.’ It was shown that neural networks that pre-learned random noise perform much faster and more accurate learning when exposed to actual data, and can achieve high learning efficiency without weight transport.
< Figure 3. Illustration depicting research on understanding the brain's operating principles through artificial neural networks >
Professor Se-Bum Paik said, “It breaks the conventional understanding of existing machine learning that only data learning is important, and provides a new perspective that focuses on the neuroscience principles of creating appropriate conditions before learning,” and added, “It is significant in that it solves important problems in artificial neural network learning through clues from developmental neuroscience, and at the same time provides insight into the brain’s learning principles through artificial neural network models.”
This study, in which Jeonghwan Cheon, a Master’s candidate of KAIST Department of Brain and Cognitive Sciences participated as the first author and Professor Sang Wan Lee of the same department as a co-author, was presented at the 38th Neural Information Processing Systems (NeurIPS), the world's top artificial intelligence conference, on December 14th in Vancouver, Canada. (Paper title: Pretraining with random noise for fast and robust learning without weight transport)
This study was conducted with the support of the National Research Foundation of Korea's Basic Research Program in Science and Engineering, the Information and Communications Technology Planning and Evaluation Institute's Talent Development Program, and the KAIST Singularity Professor Program.
2024.12.16 View 9164
KAIST Proposes a New Way to Circumvent a Long-time Frustration in Neural Computing
The human brain begins learning through spontaneous random activities even before it receives sensory information from the external world. The technology developed by the KAIST research team enables much faster and more accurate learning when exposed to actual data by pre-learning random information in a brain-mimicking artificial neural network, and is expected to be a breakthrough in the development of brain-based artificial intelligence and neuromorphic computing technology in the future.
KAIST (President Kwang-Hyung Lee) announced on the 16th of December that Professor Se-Bum Paik 's research team in the Department of Brain Cognitive Sciences solved the weight transport problem*, a long-standing challenge in neural network learning, and through this, explained the principles that enable resource-efficient learning in biological brain neural networks.
*Weight transport problem: This is the biggest obstacle to the development of artificial intelligence that mimics the biological brain. It is the fundamental reason why large-scale memory and computational work are required in the learning of general artificial neural networks, unlike biological brains.
Over the past several decades, the development of artificial intelligence has been based on error backpropagation learning proposed by Geoffery Hinton, who won the Nobel Prize in Physics this year. However, error backpropagation learning was thought to be impossible in biological brains because it requires the unrealistic assumption that individual neurons must know all the connected information across multiple layers in order to calculate the error signal for learning.
< Figure 1. Illustration depicting the method of random noise training and its effects >
This difficult problem, called the weight transport problem, was raised by Francis Crick, who won the Nobel Prize in Physiology or Medicine for the discovery of the structure of DNA, after the error backpropagation learning was proposed by Hinton in 1986. Since then, it has been considered the reason why the operating principles of natural neural networks and artificial neural networks will forever be fundamentally different.
At the borderline of artificial intelligence and neuroscience, researchers including Hinton have continued to attempt to create biologically plausible models that can implement the learning principles of the brain by solving the weight transport problem.
In 2016, a joint research team from Oxford University and DeepMind in the UK first proposed the concept of error backpropagation learning being possible without weight transport, drawing attention from the academic world. However, biologically plausible error backpropagation learning without weight transport was inefficient, with slow learning speeds and low accuracy, making it difficult to apply in reality.
KAIST research team noted that the biological brain begins learning through internal spontaneous random neural activity even before experiencing external sensory experiences. To mimic this, the research team pre-trained a biologically plausible neural network without weight transport with meaningless random information (random noise).
As a result, they showed that the symmetry of the forward and backward neural cell connections of the neural network, which is an essential condition for error backpropagation learning, can be created. In other words, learning without weight transport is possible through random pre-training.
< Figure 2. Illustration depicting the meta-learning effect of random noise training >
The research team revealed that learning random information before learning actual data has the property of meta-learning, which is ‘learning how to learn.’ It was shown that neural networks that pre-learned random noise perform much faster and more accurate learning when exposed to actual data, and can achieve high learning efficiency without weight transport.
< Figure 3. Illustration depicting research on understanding the brain's operating principles through artificial neural networks >
Professor Se-Bum Paik said, “It breaks the conventional understanding of existing machine learning that only data learning is important, and provides a new perspective that focuses on the neuroscience principles of creating appropriate conditions before learning,” and added, “It is significant in that it solves important problems in artificial neural network learning through clues from developmental neuroscience, and at the same time provides insight into the brain’s learning principles through artificial neural network models.”
This study, in which Jeonghwan Cheon, a Master’s candidate of KAIST Department of Brain and Cognitive Sciences participated as the first author and Professor Sang Wan Lee of the same department as a co-author, was presented at the 38th Neural Information Processing Systems (NeurIPS), the world's top artificial intelligence conference, on December 14th in Vancouver, Canada. (Paper title: Pretraining with random noise for fast and robust learning without weight transport)
This study was conducted with the support of the National Research Foundation of Korea's Basic Research Program in Science and Engineering, the Information and Communications Technology Planning and Evaluation Institute's Talent Development Program, and the KAIST Singularity Professor Program.
2024.12.16 View 9164 -
 KAIST Develops Janus-like Metasurface Technology that Acts According to the Direction of Light
Metasurface technology is an advanced optical technology that is thinner, lighter, and capable of precisely controlling light through nanometer-sized artificial structures compared to conventional technologies. KAIST researchers have overcome the limitations of existing metasurface technologies and successfully designed a Janus metasurface capable of perfectly controlling asymmetric light transmission. By applying this technology, they also proposed an innovative method to significantly enhance security by only decoding information under specific conditions.
KAIST (represented by President Kwang Hyung Lee) announced on the 15th of October that a research team led by Professor Jonghwa Shin from the Department of Materials Science and Engineering had developed a Janus metasurface capable of perfectly controlling asymmetric light transmission.
Asymmetric properties, which react differently depending on the direction, play a crucial role in various fields of science and engineering. The Janus metasurface developed by the research team implements an optical system capable of performing different functions in both directions.
Like the Roman god Janus with two faces, this metasurface shows entirely different optical responses depending on the direction of incoming light, effectively operating two independent optical systems with a single device (for example, a metasurface that acts as a magnifying lens in one direction and as a polarized camera in the other). In other words, by using this technology, it's possible to operate two different optical systems (e.g., a lens and a hologram) depending on the direction of the light.
This achievement addresses a challenge that existing metasurface technologies had not resolved. Conventional metasurface technology had limitations in selectively controlling the three properties of light—intensity, phase, and polarization—based on the direction of incidence.
The research team proposed a solution based on mathematical and physical principles, and succeeded in experimentally implementing different vector holograms in both directions. Through this achievement, they showcased a complete asymmetric light transmission control technology.
< Figure 1. Schematics of a device featuring asymmetric transmission. a) Device operating as a magnifying lens for back-side illumination. b) Device operating as a polarization camera for front-side illumination. >
Additionally, the research team developed a new optical encryption technology based on this metasurface technology. By using the Janus metasurface, they implemented a vector hologram that generates different images depending on the direction and polarization state of incoming light, showcasing an optical encryption system that significantly enhances security by allowing information to be decoded only under specific conditions.
This technology is expected to serve as a next-generation security solution, applicable in various fields such as quantum communication and secure data transmission.
Furthermore, the ultra-thin structure of the metasurface is expected to significantly reduce the volume and weight of traditional optical devices, contributing greatly to the miniaturization and lightweight design of next-generation devices.
< Figure 2. Experimental demonstration of Janus vectorial holograms. With front illuminations, vector images of the butterfly and the grasshopper are created, and with the back-side illuminations, vector images of the ladybug and the beetle are created. >
Professor Jonghwa Shin from the Department of Materials Science and Engineering at KAIST stated, "This research has enabled the complete asymmetric transmission control of light’s intensity, phase, and polarization, which has been a long-standing challenge in optics. It has opened up the possibility of developing various applied optical devices." He added, "We plan to continue developing optical devices that can be applied to various fields such as augmented reality (AR), holographic displays, and LiDAR systems for autonomous vehicles, utilizing the full potential of metasurface technology."
This research, in which Hyeonhee Kim (a doctoral student in the Department of Materials Science and Engineering at KAIST) and Joonkyo Jung participated as co-first authors, was published online in the international journal Advanced Materials and is scheduled to be published in the October 31 issue. (Title of the paper: "Bidirectional Vectorial Holography Using Bi-Layer Metasurfaces and Its Application to Optical Encryption")
The research was supported by the Nano Materials Technology Development Program and the Mid-Career Researcher Program of the National Research Foundation of Korea.
2024.10.15 View 5134
KAIST Develops Janus-like Metasurface Technology that Acts According to the Direction of Light
Metasurface technology is an advanced optical technology that is thinner, lighter, and capable of precisely controlling light through nanometer-sized artificial structures compared to conventional technologies. KAIST researchers have overcome the limitations of existing metasurface technologies and successfully designed a Janus metasurface capable of perfectly controlling asymmetric light transmission. By applying this technology, they also proposed an innovative method to significantly enhance security by only decoding information under specific conditions.
KAIST (represented by President Kwang Hyung Lee) announced on the 15th of October that a research team led by Professor Jonghwa Shin from the Department of Materials Science and Engineering had developed a Janus metasurface capable of perfectly controlling asymmetric light transmission.
Asymmetric properties, which react differently depending on the direction, play a crucial role in various fields of science and engineering. The Janus metasurface developed by the research team implements an optical system capable of performing different functions in both directions.
Like the Roman god Janus with two faces, this metasurface shows entirely different optical responses depending on the direction of incoming light, effectively operating two independent optical systems with a single device (for example, a metasurface that acts as a magnifying lens in one direction and as a polarized camera in the other). In other words, by using this technology, it's possible to operate two different optical systems (e.g., a lens and a hologram) depending on the direction of the light.
This achievement addresses a challenge that existing metasurface technologies had not resolved. Conventional metasurface technology had limitations in selectively controlling the three properties of light—intensity, phase, and polarization—based on the direction of incidence.
The research team proposed a solution based on mathematical and physical principles, and succeeded in experimentally implementing different vector holograms in both directions. Through this achievement, they showcased a complete asymmetric light transmission control technology.
< Figure 1. Schematics of a device featuring asymmetric transmission. a) Device operating as a magnifying lens for back-side illumination. b) Device operating as a polarization camera for front-side illumination. >
Additionally, the research team developed a new optical encryption technology based on this metasurface technology. By using the Janus metasurface, they implemented a vector hologram that generates different images depending on the direction and polarization state of incoming light, showcasing an optical encryption system that significantly enhances security by allowing information to be decoded only under specific conditions.
This technology is expected to serve as a next-generation security solution, applicable in various fields such as quantum communication and secure data transmission.
Furthermore, the ultra-thin structure of the metasurface is expected to significantly reduce the volume and weight of traditional optical devices, contributing greatly to the miniaturization and lightweight design of next-generation devices.
< Figure 2. Experimental demonstration of Janus vectorial holograms. With front illuminations, vector images of the butterfly and the grasshopper are created, and with the back-side illuminations, vector images of the ladybug and the beetle are created. >
Professor Jonghwa Shin from the Department of Materials Science and Engineering at KAIST stated, "This research has enabled the complete asymmetric transmission control of light’s intensity, phase, and polarization, which has been a long-standing challenge in optics. It has opened up the possibility of developing various applied optical devices." He added, "We plan to continue developing optical devices that can be applied to various fields such as augmented reality (AR), holographic displays, and LiDAR systems for autonomous vehicles, utilizing the full potential of metasurface technology."
This research, in which Hyeonhee Kim (a doctoral student in the Department of Materials Science and Engineering at KAIST) and Joonkyo Jung participated as co-first authors, was published online in the international journal Advanced Materials and is scheduled to be published in the October 31 issue. (Title of the paper: "Bidirectional Vectorial Holography Using Bi-Layer Metasurfaces and Its Application to Optical Encryption")
The research was supported by the Nano Materials Technology Development Program and the Mid-Career Researcher Program of the National Research Foundation of Korea.
2024.10.15 View 5134