bio
-
 A New Therapeutic Drug for Alzheimer’s Disease without Inflammatory Side Effects
Although Aduhelm, a monoclonal antibody targeting amyloid beta (Aβ), recently became the first US FDA approved drug for Alzheimer’s disease (AD) based on its ability to decrease Aβ plaque burden in AD patients, its effect on cognitive improvement is still controversial. Moreover, about 40% of the patients treated with this antibody experienced serious side effects including cerebral edemas (ARIA-E) and hemorrhages (ARIA-H) that are likely related to inflammatory responses in the brain when the Aβ antibody binds Fc receptors (FCR) of immune cells such as microglia and macrophages.
These inflammatory side effects can cause neuronal cell death and synapse elimination by activated microglia, and even have the potential to exacerbate cognitive impairment in AD patients. Thus, current Aβ antibody-based immunotherapy holds the inherent risk of doing more harm than good due to their inflammatory side effects.
To overcome these problems, a team of researchers at KAIST in South Korea has developed a novel fusion protein drug, αAβ-Gas6, which efficiently eliminates Aβ via an entirely different mechanism than Aβ antibody-based immunotherapy. In a mouse model of AD, αAβ-Gas6 not only removed Aβ with higher potency, but also circumvented the neurotoxic inflammatory side effects associated with conventional antibody treatments.
Their findings were published on August 4 in Nature Medicine.
Schematic of a chimeric Gas6 fusion protein. A single chain variable fragment (scFv) of
an Amyloid β (Aβ)-targeting monoclonal antibody is fused with a truncated receptor binding
domain of Gas6, a bridging molecule for the clearance of dead cells via TAM (TYRO3, AXL,
and MERTK) receptors, which are expressed by microglia and astrocytes.
“FcR activation by Aβ targeting antibodies induces microglia-mediated Aβ phagocytosis, but it also produces inflammatory signals, inevitably damaging brain tissues,” said paper authors Chan Hyuk Kim and Won-Suk Chung, associate professors in the Department of Biological Sciences at KAIST.
“Therefore, we utilized efferocytosis, a cellular process by which dead cells are removed by phagocytes as an alternative pathway for the clearance of Aβ in the brain,” Prof. Kim and Chung said. “Efferocytosis is accompanied by anti-inflammatory responses to maintain tissue homeostasis. To exploit this process, we engineered Gas6, a soluble adaptor protein that mediates efferocytosis via TAM phagocytic receptors in such a way that its target specificity was redirected from dead cells to Aβ plaques.”
The professors and their team demonstrated that the resulting αAβ-Gas6 induced Aβ engulfment by activating not only microglial but also astrocytic phagocytosis since TAM phagocytic receptors are highly expressed by these two major phagocytes in the brain. Importantly, αAβ-Gas6 promoted the robust uptake of Aβ without showing any signs of inflammation and neurotoxicity, which contrasts sharply with the treatment using an Aβ monoclonal antibody. Moreover, they showed that αAβ-Gas6 substantially reduced excessive synapse elimination by microglia, consequently leading to better behavioral rescues in AD model mice.
“By using a mouse model of cerebral amyloid angiopathy (CAA), a cerebrovascular disorder caused by the deposition of Aβ within the walls of the brain’s blood vessels, we also showed that the intrathecal administration of Gas6 fusion protein significantly eliminated cerebrovascular amyloids, along with a reduction of microhemorrhages. These data demonstrate that aAb-Gas6 is a potent therapeutic agent in eliminating Aβ without exacerbating CAA-related microhemorrhages.”
The resulting αAβ-Gas6 clears Aβ oligomers and fibrils without causing neurotoxicity (a-b, neurons: red, and fragmented axons: yellow) and proinflammatory responses (c, TNF release), which are conversely exacerbated by the treatment of an Aβ-targeting monoclonal antibody (Aducanumab).
Professors Kim and Chung noted, “We believe our approach can be a breakthrough in treating AD without causing inflammatory side effects and synapse loss. Our approach holds promise as a novel therapeutic platform that is applicable to more than AD. By modifying the target-specificity of the fusion protein, the Gas6-fusion protein can be applied to various neurological disorders as well as autoimmune diseases affected by toxic molecules that should be removed without causing inflammatory responses.”
The number and total area of Aβ plaques (Thioflavin-T, green) were significantly reduced in αAβ-Gas6-treated AD mouse brains compared to Aducanumab-treated ones (a, b). The cognitive functions of AD model mice were significantly rescued by αAβ-Gas6 treatment, whereas Aducanumab-treated AD mice showed partial rescue in these cognitive tests (c-e).
Professors Kim and Chung founded “Illimis Therapeutics” based on this strategy of designing chimeric Gas6 fusion proteins that would remove toxic aggregates from the nervous system. Through this company, they are planning to further develop various Gas6-fusion proteins not only for Ab but also for Tau to treat AD symptoms.
This work was supported by KAIST and the Korea Health Technology R&D Project that was administered by the Korea Health Industry Development Institute (KHIDI) and the Korea Dementia Research Center (KDRC) funded by the Ministry of Health & Welfare (MOHW) and the Ministry of Science and ICT (MSIT), and KAIST.
Other contributors include Hyuncheol Jung and Se Young Lee, Sungjoon Lim, Hyeong Ryeol Choi, Yeseong Choi, Minjin Kim, Segi Kim, the Department of Biological Sciences, and the Korea Advanced Institute of Science and Technology (KAIST).
To receive more up-to-date information on this new development, follow “Illimis Therapeutics” on twitter @Illimistx.
2022.08.05 View 14866
A New Therapeutic Drug for Alzheimer’s Disease without Inflammatory Side Effects
Although Aduhelm, a monoclonal antibody targeting amyloid beta (Aβ), recently became the first US FDA approved drug for Alzheimer’s disease (AD) based on its ability to decrease Aβ plaque burden in AD patients, its effect on cognitive improvement is still controversial. Moreover, about 40% of the patients treated with this antibody experienced serious side effects including cerebral edemas (ARIA-E) and hemorrhages (ARIA-H) that are likely related to inflammatory responses in the brain when the Aβ antibody binds Fc receptors (FCR) of immune cells such as microglia and macrophages.
These inflammatory side effects can cause neuronal cell death and synapse elimination by activated microglia, and even have the potential to exacerbate cognitive impairment in AD patients. Thus, current Aβ antibody-based immunotherapy holds the inherent risk of doing more harm than good due to their inflammatory side effects.
To overcome these problems, a team of researchers at KAIST in South Korea has developed a novel fusion protein drug, αAβ-Gas6, which efficiently eliminates Aβ via an entirely different mechanism than Aβ antibody-based immunotherapy. In a mouse model of AD, αAβ-Gas6 not only removed Aβ with higher potency, but also circumvented the neurotoxic inflammatory side effects associated with conventional antibody treatments.
Their findings were published on August 4 in Nature Medicine.
Schematic of a chimeric Gas6 fusion protein. A single chain variable fragment (scFv) of
an Amyloid β (Aβ)-targeting monoclonal antibody is fused with a truncated receptor binding
domain of Gas6, a bridging molecule for the clearance of dead cells via TAM (TYRO3, AXL,
and MERTK) receptors, which are expressed by microglia and astrocytes.
“FcR activation by Aβ targeting antibodies induces microglia-mediated Aβ phagocytosis, but it also produces inflammatory signals, inevitably damaging brain tissues,” said paper authors Chan Hyuk Kim and Won-Suk Chung, associate professors in the Department of Biological Sciences at KAIST.
“Therefore, we utilized efferocytosis, a cellular process by which dead cells are removed by phagocytes as an alternative pathway for the clearance of Aβ in the brain,” Prof. Kim and Chung said. “Efferocytosis is accompanied by anti-inflammatory responses to maintain tissue homeostasis. To exploit this process, we engineered Gas6, a soluble adaptor protein that mediates efferocytosis via TAM phagocytic receptors in such a way that its target specificity was redirected from dead cells to Aβ plaques.”
The professors and their team demonstrated that the resulting αAβ-Gas6 induced Aβ engulfment by activating not only microglial but also astrocytic phagocytosis since TAM phagocytic receptors are highly expressed by these two major phagocytes in the brain. Importantly, αAβ-Gas6 promoted the robust uptake of Aβ without showing any signs of inflammation and neurotoxicity, which contrasts sharply with the treatment using an Aβ monoclonal antibody. Moreover, they showed that αAβ-Gas6 substantially reduced excessive synapse elimination by microglia, consequently leading to better behavioral rescues in AD model mice.
“By using a mouse model of cerebral amyloid angiopathy (CAA), a cerebrovascular disorder caused by the deposition of Aβ within the walls of the brain’s blood vessels, we also showed that the intrathecal administration of Gas6 fusion protein significantly eliminated cerebrovascular amyloids, along with a reduction of microhemorrhages. These data demonstrate that aAb-Gas6 is a potent therapeutic agent in eliminating Aβ without exacerbating CAA-related microhemorrhages.”
The resulting αAβ-Gas6 clears Aβ oligomers and fibrils without causing neurotoxicity (a-b, neurons: red, and fragmented axons: yellow) and proinflammatory responses (c, TNF release), which are conversely exacerbated by the treatment of an Aβ-targeting monoclonal antibody (Aducanumab).
Professors Kim and Chung noted, “We believe our approach can be a breakthrough in treating AD without causing inflammatory side effects and synapse loss. Our approach holds promise as a novel therapeutic platform that is applicable to more than AD. By modifying the target-specificity of the fusion protein, the Gas6-fusion protein can be applied to various neurological disorders as well as autoimmune diseases affected by toxic molecules that should be removed without causing inflammatory responses.”
The number and total area of Aβ plaques (Thioflavin-T, green) were significantly reduced in αAβ-Gas6-treated AD mouse brains compared to Aducanumab-treated ones (a, b). The cognitive functions of AD model mice were significantly rescued by αAβ-Gas6 treatment, whereas Aducanumab-treated AD mice showed partial rescue in these cognitive tests (c-e).
Professors Kim and Chung founded “Illimis Therapeutics” based on this strategy of designing chimeric Gas6 fusion proteins that would remove toxic aggregates from the nervous system. Through this company, they are planning to further develop various Gas6-fusion proteins not only for Ab but also for Tau to treat AD symptoms.
This work was supported by KAIST and the Korea Health Technology R&D Project that was administered by the Korea Health Industry Development Institute (KHIDI) and the Korea Dementia Research Center (KDRC) funded by the Ministry of Health & Welfare (MOHW) and the Ministry of Science and ICT (MSIT), and KAIST.
Other contributors include Hyuncheol Jung and Se Young Lee, Sungjoon Lim, Hyeong Ryeol Choi, Yeseong Choi, Minjin Kim, Segi Kim, the Department of Biological Sciences, and the Korea Advanced Institute of Science and Technology (KAIST).
To receive more up-to-date information on this new development, follow “Illimis Therapeutics” on twitter @Illimistx.
2022.08.05 View 14866 -
 Metabolically Engineered Bacterium Produces Lutein
A research group at KAIST has engineered a bacterial strain capable of producing lutein. The research team applied systems metabolic engineering strategies, including substrate channeling and electron channeling, to enhance the production of lutein in an engineered Escherichia coli strain. The strategies will be also useful for the efficient production of other industrially important natural products used in the food, pharmaceutical, and cosmetic industries.
Figure: Systems metabolic engineering was employed to construct and optimize the metabolic pathways for lutein production, and substrate channeling and electron channeling strategies were additionally employed to increase the production of the lutein with high productivity.
Lutein is classified as a xanthophyll chemical that is abundant in egg yolk, fruits, and vegetables. It protects the eye from oxidative damage from radiation and reduces the risk of eye diseases including macular degeneration and cataracts. Commercialized products featuring lutein are derived from the extracts of the marigold flower, which is known to harbor abundant amounts of lutein. However, the drawback of lutein production from nature is that it takes a long time to grow and harvest marigold flowers. Furthermore, it requires additional physical and chemical-based extractions with a low yield, which makes it economically unfeasible in terms of productivity. The high cost and low yield of these bioprocesses has made it difficult to readily meet the demand for lutein.
These challenges inspired the metabolic engineers at KAIST, including researchers Dr. Seon Young Park, Ph.D. Candidate Hyunmin Eun, and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering. The team’s study entitled “Metabolic engineering of Escherichia coli with electron channeling for the production of natural products” was published in Nature Catalysis on August 5, 2022.
This research details the ability to produce lutein from E. coli with a high yield using a cheap carbon source, glycerol, via systems metabolic engineering. The research group focused on solving the bottlenecks of the biosynthetic pathway for lutein production constructed within an individual cell. First, using systems metabolic engineering, which is an integrated technology to engineer the metabolism of a microorganism, lutein was produced when the lutein biosynthesis pathway was introduced, albeit in very small amounts.
To improve the productivity of lutein production, the bottleneck enzymes within the metabolic pathway were first identified. It turned out that metabolic reactions that involve a promiscuous enzyme, an enzyme that is involved in two or more metabolic reactions, and electron-requiring cytochrome P450 enzymes are the main bottleneck steps of the pathway inhibiting lutein biosynthesis.
To overcome these challenges, substrate channeling, a strategy to artificially recruit enzymes in physical proximity within the cell in order to increase the local concentrations of substrates that can be converted into products, was employed to channel more metabolic flux towards the target chemical while reducing the formation of unwanted byproducts.
Furthermore, electron channeling, a strategy similar to substrate channeling but differing in terms of increasing the local concentrations of electrons required for oxidoreduction reactions mediated by P450 and its reductase partners, was applied to further streamline the metabolic flux towards lutein biosynthesis, which led to the highest titer of lutein production achieved in a bacterial host ever reported. The same electron channeling strategy was successfully applied for the production of other natural products including nootkatone and apigenin in E. coli, showcasing the general applicability of the strategy in the research field.
“It is expected that this microbial cell factory-based production of lutein will be able to replace the current plant extraction-based process,” said Dr. Seon Young Park, the first author of the paper. She explained that another important point of the research is that integrated metabolic engineering strategies developed from this study can be generally applicable for the efficient production of other natural products useful as pharmaceuticals or nutraceuticals.
“As maintaining good health in an aging society is becoming increasingly important, we expect that the technology and strategies developed here will play pivotal roles in producing other valuable natural products of medical or nutritional importance,” explained Distinguished Professor Sang Yup Lee.
This work was supported by the Cooperative Research Program for Agriculture Science & Technology Development funded by the Rural Development Administration of Korea, with further support from the Development of Next-generation Biorefinery Platform Technologies for Leading Bio-based Chemicals Industry Project and by the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project of the National Research Foundation funded by the Ministry of Science and ICT of Korea.
2022.08.05 View 12486
Metabolically Engineered Bacterium Produces Lutein
A research group at KAIST has engineered a bacterial strain capable of producing lutein. The research team applied systems metabolic engineering strategies, including substrate channeling and electron channeling, to enhance the production of lutein in an engineered Escherichia coli strain. The strategies will be also useful for the efficient production of other industrially important natural products used in the food, pharmaceutical, and cosmetic industries.
Figure: Systems metabolic engineering was employed to construct and optimize the metabolic pathways for lutein production, and substrate channeling and electron channeling strategies were additionally employed to increase the production of the lutein with high productivity.
Lutein is classified as a xanthophyll chemical that is abundant in egg yolk, fruits, and vegetables. It protects the eye from oxidative damage from radiation and reduces the risk of eye diseases including macular degeneration and cataracts. Commercialized products featuring lutein are derived from the extracts of the marigold flower, which is known to harbor abundant amounts of lutein. However, the drawback of lutein production from nature is that it takes a long time to grow and harvest marigold flowers. Furthermore, it requires additional physical and chemical-based extractions with a low yield, which makes it economically unfeasible in terms of productivity. The high cost and low yield of these bioprocesses has made it difficult to readily meet the demand for lutein.
These challenges inspired the metabolic engineers at KAIST, including researchers Dr. Seon Young Park, Ph.D. Candidate Hyunmin Eun, and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering. The team’s study entitled “Metabolic engineering of Escherichia coli with electron channeling for the production of natural products” was published in Nature Catalysis on August 5, 2022.
This research details the ability to produce lutein from E. coli with a high yield using a cheap carbon source, glycerol, via systems metabolic engineering. The research group focused on solving the bottlenecks of the biosynthetic pathway for lutein production constructed within an individual cell. First, using systems metabolic engineering, which is an integrated technology to engineer the metabolism of a microorganism, lutein was produced when the lutein biosynthesis pathway was introduced, albeit in very small amounts.
To improve the productivity of lutein production, the bottleneck enzymes within the metabolic pathway were first identified. It turned out that metabolic reactions that involve a promiscuous enzyme, an enzyme that is involved in two or more metabolic reactions, and electron-requiring cytochrome P450 enzymes are the main bottleneck steps of the pathway inhibiting lutein biosynthesis.
To overcome these challenges, substrate channeling, a strategy to artificially recruit enzymes in physical proximity within the cell in order to increase the local concentrations of substrates that can be converted into products, was employed to channel more metabolic flux towards the target chemical while reducing the formation of unwanted byproducts.
Furthermore, electron channeling, a strategy similar to substrate channeling but differing in terms of increasing the local concentrations of electrons required for oxidoreduction reactions mediated by P450 and its reductase partners, was applied to further streamline the metabolic flux towards lutein biosynthesis, which led to the highest titer of lutein production achieved in a bacterial host ever reported. The same electron channeling strategy was successfully applied for the production of other natural products including nootkatone and apigenin in E. coli, showcasing the general applicability of the strategy in the research field.
“It is expected that this microbial cell factory-based production of lutein will be able to replace the current plant extraction-based process,” said Dr. Seon Young Park, the first author of the paper. She explained that another important point of the research is that integrated metabolic engineering strategies developed from this study can be generally applicable for the efficient production of other natural products useful as pharmaceuticals or nutraceuticals.
“As maintaining good health in an aging society is becoming increasingly important, we expect that the technology and strategies developed here will play pivotal roles in producing other valuable natural products of medical or nutritional importance,” explained Distinguished Professor Sang Yup Lee.
This work was supported by the Cooperative Research Program for Agriculture Science & Technology Development funded by the Rural Development Administration of Korea, with further support from the Development of Next-generation Biorefinery Platform Technologies for Leading Bio-based Chemicals Industry Project and by the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project of the National Research Foundation funded by the Ministry of Science and ICT of Korea.
2022.08.05 View 12486 -
 VP Sang Yup Lee Receives Honorary Doctorate from DTU
Vice President for Research, Distinguished Professor Sang Yup Lee at the Department of Chemical & Biomolecular Engineering, was awarded an honorary doctorate from the Technical University of Denmark (DTU) during the DTU Commemoration Day 2022 on April 29. The event drew distinguished guests, students, and faculty including HRH The Crown Prince Frederik Andre Henrik Christian and DTU President Anders Bjarklev.
Professor Lee was recognized for his exceptional scholarship in the field of systems metabolic engineering, which led to the development of microcell factories capable of producing a wide range of fuels, chemicals, materials, and natural compounds, many for the first time.
Professor Lee said in his acceptance speech that KAIST’s continued partnership with DTU in the field of biotechnology will lead to significant contributions in the global efforts to respond to climate change and promote green growth.
DTU CPO and CSO Dina Petronovic Nielson, who heads DTU Biosustain, also lauded Professor Lee saying, “It is not only a great honor for Professor Lee to be induced at DTU but also great honor for DTU to have him.”
Professor Lee also gave commemorative lectures at DTU Biosustain in Lingby and the Bio Innovation Research Institute at the Novo Nordisk Foundation in Copenhagen while in Denmark.
DTU, one of the leading science and technology universities in Europe, has been awarding honorary doctorates since 1921, including to Nobel laureate in chemistry Professor Frances Arnold at Caltech. Professor Lee is the first Korean to receive an honorary doctorate from DTU.
2022.05.03 View 13341
VP Sang Yup Lee Receives Honorary Doctorate from DTU
Vice President for Research, Distinguished Professor Sang Yup Lee at the Department of Chemical & Biomolecular Engineering, was awarded an honorary doctorate from the Technical University of Denmark (DTU) during the DTU Commemoration Day 2022 on April 29. The event drew distinguished guests, students, and faculty including HRH The Crown Prince Frederik Andre Henrik Christian and DTU President Anders Bjarklev.
Professor Lee was recognized for his exceptional scholarship in the field of systems metabolic engineering, which led to the development of microcell factories capable of producing a wide range of fuels, chemicals, materials, and natural compounds, many for the first time.
Professor Lee said in his acceptance speech that KAIST’s continued partnership with DTU in the field of biotechnology will lead to significant contributions in the global efforts to respond to climate change and promote green growth.
DTU CPO and CSO Dina Petronovic Nielson, who heads DTU Biosustain, also lauded Professor Lee saying, “It is not only a great honor for Professor Lee to be induced at DTU but also great honor for DTU to have him.”
Professor Lee also gave commemorative lectures at DTU Biosustain in Lingby and the Bio Innovation Research Institute at the Novo Nordisk Foundation in Copenhagen while in Denmark.
DTU, one of the leading science and technology universities in Europe, has been awarding honorary doctorates since 1921, including to Nobel laureate in chemistry Professor Frances Arnold at Caltech. Professor Lee is the first Korean to receive an honorary doctorate from DTU.
2022.05.03 View 13341 -
 Decoding Brain Signals to Control a Robotic Arm
Advanced brain-machine interface system successfully interprets arm movement directions from neural signals in the brain
Researchers have developed a mind-reading system for decoding neural signals from the brain during arm movement. The method, described in the journal Applied Soft Computing, can be used by a person to control a robotic arm through a brain-machine interface (BMI).
A BMI is a device that translates nerve signals into commands to control a machine, such as a computer or a robotic limb. There are two main techniques for monitoring neural signals in BMIs: electroencephalography (EEG) and electrocorticography (ECoG).
The EEG exhibits signals from electrodes on the surface of the scalp and is widely employed because it is non-invasive, relatively cheap, safe and easy to use. However, the EEG has low spatial resolution and detects irrelevant neural signals, which makes it difficult to interpret the intentions of individuals from the EEG.
On the other hand, the ECoG is an invasive method that involves placing electrodes directly on the surface of the cerebral cortex below the scalp. Compared with the EEG, the ECoG can monitor neural signals with much higher spatial resolution and less background noise. However, this technique has several drawbacks.
“The ECoG is primarily used to find potential sources of epileptic seizures, meaning the electrodes are placed in different locations for different patients and may not be in the optimal regions of the brain for detecting sensory and movement signals,” explained Professor Jaeseung Jeong, a brain scientist at KAIST. “This inconsistency makes it difficult to decode brain signals to predict movements.”
To overcome these problems, Professor Jeong’s team developed a new method for decoding ECoG neural signals during arm movement. The system is based on a machine-learning system for analysing and predicting neural signals called an ‘echo-state network’ and a mathematical probability model called the Gaussian distribution.
In the study, the researchers recorded ECoG signals from four individuals with epilepsy while they were performing a reach-and-grasp task. Because the ECoG electrodes were placed according to the potential sources of each patient’s epileptic seizures, only 22% to 44% of the electrodes were located in the regions of the brain responsible for controlling movement.
During the movement task, the participants were given visual cues, either by placing a real tennis ball in front of them, or via a virtual reality headset showing a clip of a human arm reaching forward in first-person view. They were asked to reach forward, grasp an object, then return their hand and release the object, while wearing motion sensors on their wrists and fingers. In a second task, they were instructed to imagine reaching forward without moving their arms.
The researchers monitored the signals from the ECoG electrodes during real and imaginary arm movements, and tested whether the new system could predict the direction of this movement from the neural signals. They found that the novel decoder successfully classified arm movements in 24 directions in three-dimensional space, both in the real and virtual tasks, and that the results were at least five times more accurate than chance. They also used a computer simulation to show that the novel ECoG decoder could control the movements of a robotic arm.
Overall, the results suggest that the new machine learning-based BCI system successfully used ECoG signals to interpret the direction of the intended movements. The next steps will be to improve the accuracy and efficiency of the decoder. In the future, it could be used in a real-time BMI device to help people with movement or sensory impairments.
This research was supported by the KAIST Global Singularity Research Program of 2021, Brain Research Program of the National Research Foundation of Korea funded by the Ministry of Science, ICT, and Future Planning, and the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education.
-PublicationHoon-Hee Kim, Jaeseung Jeong, “An electrocorticographic decoder for arm movement for brain-machine interface using an echo state network and Gaussian readout,” Applied SoftComputing online December 31, 2021 (doi.org/10.1016/j.asoc.2021.108393)
-ProfileProfessor Jaeseung JeongDepartment of Bio and Brain EngineeringCollege of EngineeringKAIST
2022.03.18 View 14916
Decoding Brain Signals to Control a Robotic Arm
Advanced brain-machine interface system successfully interprets arm movement directions from neural signals in the brain
Researchers have developed a mind-reading system for decoding neural signals from the brain during arm movement. The method, described in the journal Applied Soft Computing, can be used by a person to control a robotic arm through a brain-machine interface (BMI).
A BMI is a device that translates nerve signals into commands to control a machine, such as a computer or a robotic limb. There are two main techniques for monitoring neural signals in BMIs: electroencephalography (EEG) and electrocorticography (ECoG).
The EEG exhibits signals from electrodes on the surface of the scalp and is widely employed because it is non-invasive, relatively cheap, safe and easy to use. However, the EEG has low spatial resolution and detects irrelevant neural signals, which makes it difficult to interpret the intentions of individuals from the EEG.
On the other hand, the ECoG is an invasive method that involves placing electrodes directly on the surface of the cerebral cortex below the scalp. Compared with the EEG, the ECoG can monitor neural signals with much higher spatial resolution and less background noise. However, this technique has several drawbacks.
“The ECoG is primarily used to find potential sources of epileptic seizures, meaning the electrodes are placed in different locations for different patients and may not be in the optimal regions of the brain for detecting sensory and movement signals,” explained Professor Jaeseung Jeong, a brain scientist at KAIST. “This inconsistency makes it difficult to decode brain signals to predict movements.”
To overcome these problems, Professor Jeong’s team developed a new method for decoding ECoG neural signals during arm movement. The system is based on a machine-learning system for analysing and predicting neural signals called an ‘echo-state network’ and a mathematical probability model called the Gaussian distribution.
In the study, the researchers recorded ECoG signals from four individuals with epilepsy while they were performing a reach-and-grasp task. Because the ECoG electrodes were placed according to the potential sources of each patient’s epileptic seizures, only 22% to 44% of the electrodes were located in the regions of the brain responsible for controlling movement.
During the movement task, the participants were given visual cues, either by placing a real tennis ball in front of them, or via a virtual reality headset showing a clip of a human arm reaching forward in first-person view. They were asked to reach forward, grasp an object, then return their hand and release the object, while wearing motion sensors on their wrists and fingers. In a second task, they were instructed to imagine reaching forward without moving their arms.
The researchers monitored the signals from the ECoG electrodes during real and imaginary arm movements, and tested whether the new system could predict the direction of this movement from the neural signals. They found that the novel decoder successfully classified arm movements in 24 directions in three-dimensional space, both in the real and virtual tasks, and that the results were at least five times more accurate than chance. They also used a computer simulation to show that the novel ECoG decoder could control the movements of a robotic arm.
Overall, the results suggest that the new machine learning-based BCI system successfully used ECoG signals to interpret the direction of the intended movements. The next steps will be to improve the accuracy and efficiency of the decoder. In the future, it could be used in a real-time BMI device to help people with movement or sensory impairments.
This research was supported by the KAIST Global Singularity Research Program of 2021, Brain Research Program of the National Research Foundation of Korea funded by the Ministry of Science, ICT, and Future Planning, and the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education.
-PublicationHoon-Hee Kim, Jaeseung Jeong, “An electrocorticographic decoder for arm movement for brain-machine interface using an echo state network and Gaussian readout,” Applied SoftComputing online December 31, 2021 (doi.org/10.1016/j.asoc.2021.108393)
-ProfileProfessor Jaeseung JeongDepartment of Bio and Brain EngineeringCollege of EngineeringKAIST
2022.03.18 View 14916 -
 Five Projects Ranked in the Top 100 for National R&D Excellence
Five KAIST research projects were selected as the 2021 Top 100 for National R&D Excellence by the Ministry of Science and ICT and the Korea Institute of Science & Technology Evaluation and Planning.
The five projects are:-The development of E. coli that proliferates with only formic acid and carbon dioxide by Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering
-An original reverse aging technology that restores an old human skin cell into a younger one by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering-The development of next-generation high-efficiency perovskite-silicon tandem solar cells by Professor Byungha Shin from the Department of Materials Science and Engineering-Research on the effects of ultrafine dust in the atmosphere has on energy consumption by Professor Jiyong Eom from the School of Business and Technology Management-Research on a molecular trigger that controls the phase transformation of bio materials by Professor Myungchul Kim from the Department of Bio and Brain Engineering
Started in 2006, an Evaluation Committee composed of experts in industries, universities, and research institutes has made the preliminary selections of the most outstanding research projects based on their significance as a scientific and technological development and their socioeconomic effects. The finalists went through an open public evaluation. The final 100 studies are from six fields: 18 from mechanics & materials, 26 from biology & marine sciences, 19 from ICT & electronics, 10 from interdisciplinary research, and nine from natural science and infrastructure.
The selected 100 studies will receive a certificate and an award plaque from the minister of MSIT as well as additional points for business and institutional evaluations according to appropriate regulations, and the selected researchers will be strongly recommended as candidates for national meritorious awards.
In particular, to help the 100 selected research projects become more accessible for the general public, their main contents will be provided in a free e-book ‘The Top 100 for National R&D Excellence of 2021’ that will be available from online booksellers.
2022.02.17 View 12986
Five Projects Ranked in the Top 100 for National R&D Excellence
Five KAIST research projects were selected as the 2021 Top 100 for National R&D Excellence by the Ministry of Science and ICT and the Korea Institute of Science & Technology Evaluation and Planning.
The five projects are:-The development of E. coli that proliferates with only formic acid and carbon dioxide by Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering
-An original reverse aging technology that restores an old human skin cell into a younger one by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering-The development of next-generation high-efficiency perovskite-silicon tandem solar cells by Professor Byungha Shin from the Department of Materials Science and Engineering-Research on the effects of ultrafine dust in the atmosphere has on energy consumption by Professor Jiyong Eom from the School of Business and Technology Management-Research on a molecular trigger that controls the phase transformation of bio materials by Professor Myungchul Kim from the Department of Bio and Brain Engineering
Started in 2006, an Evaluation Committee composed of experts in industries, universities, and research institutes has made the preliminary selections of the most outstanding research projects based on their significance as a scientific and technological development and their socioeconomic effects. The finalists went through an open public evaluation. The final 100 studies are from six fields: 18 from mechanics & materials, 26 from biology & marine sciences, 19 from ICT & electronics, 10 from interdisciplinary research, and nine from natural science and infrastructure.
The selected 100 studies will receive a certificate and an award plaque from the minister of MSIT as well as additional points for business and institutional evaluations according to appropriate regulations, and the selected researchers will be strongly recommended as candidates for national meritorious awards.
In particular, to help the 100 selected research projects become more accessible for the general public, their main contents will be provided in a free e-book ‘The Top 100 for National R&D Excellence of 2021’ that will be available from online booksellers.
2022.02.17 View 12986 -
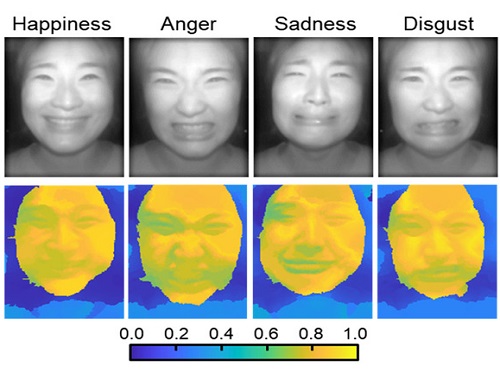 AI Light-Field Camera Reads 3D Facial Expressions
Machine-learned, light-field camera reads facial expressions from high-contrast illumination invariant 3D facial images
A joint research team led by Professors Ki-Hun Jeong and Doheon Lee from the KAIST Department of Bio and Brain Engineering reported the development of a technique for facial expression detection by merging near-infrared light-field camera techniques with artificial intelligence (AI) technology.
Unlike a conventional camera, the light-field camera contains micro-lens arrays in front of the image sensor, which makes the camera small enough to fit into a smart phone, while allowing it to acquire the spatial and directional information of the light with a single shot. The technique has received attention as it can reconstruct images in a variety of ways including multi-views, refocusing, and 3D image acquisition, giving rise to many potential applications.
However, the optical crosstalk between shadows caused by external light sources in the environment and the micro-lens has limited existing light-field cameras from being able to provide accurate image contrast and 3D reconstruction.
The joint research team applied a vertical-cavity surface-emitting laser (VCSEL) in the near-IR range to stabilize the accuracy of 3D image reconstruction that previously depended on environmental light. When an external light source is shone on a face at 0-, 30-, and 60-degree angles, the light field camera reduces 54% of image reconstruction errors. Additionally, by inserting a light-absorbing layer for visible and near-IR wavelengths between the micro-lens arrays, the team could minimize optical crosstalk while increasing the image contrast by 2.1 times.
Through this technique, the team could overcome the limitations of existing light-field cameras and was able to develop their NIR-based light-field camera (NIR-LFC), optimized for the 3D image reconstruction of facial expressions. Using the NIR-LFC, the team acquired high-quality 3D reconstruction images of facial expressions expressing various emotions regardless of the lighting conditions of the surrounding environment.
The facial expressions in the acquired 3D images were distinguished through machine learning with an average of 85% accuracy – a statistically significant figure compared to when 2D images were used. Furthermore, by calculating the interdependency of distance information that varies with facial expression in 3D images, the team could identify the information a light-field camera utilizes to distinguish human expressions.
Professor Ki-Hun Jeong said, “The sub-miniature light-field camera developed by the research team has the potential to become the new platform to quantitatively analyze the facial expressions and emotions of humans.” To highlight the significance of this research, he added, “It could be applied in various fields including mobile healthcare, field diagnosis, social cognition, and human-machine interactions.”
This research was published in Advanced Intelligent Systems online on December 16, under the title, “Machine-Learned Light-field Camera that Reads Facial Expression from High-Contrast and Illumination Invariant 3D Facial Images.” This research was funded by the Ministry of Science and ICT and the Ministry of Trade, Industry and Energy.
-Publication“Machine-learned light-field camera that reads fascial expression from high-contrast and illumination invariant 3D facial images,” Sang-In Bae, Sangyeon Lee, Jae-Myeong Kwon, Hyun-Kyung Kim. Kyung-Won Jang, Doheon Lee, Ki-Hun Jeong, Advanced Intelligent Systems, December 16, 2021 (doi.org/10.1002/aisy.202100182)
ProfileProfessor Ki-Hun JeongBiophotonic LaboratoryDepartment of Bio and Brain EngineeringKAIST
Professor Doheon LeeDepartment of Bio and Brain EngineeringKAIST
2022.01.21 View 15030
AI Light-Field Camera Reads 3D Facial Expressions
Machine-learned, light-field camera reads facial expressions from high-contrast illumination invariant 3D facial images
A joint research team led by Professors Ki-Hun Jeong and Doheon Lee from the KAIST Department of Bio and Brain Engineering reported the development of a technique for facial expression detection by merging near-infrared light-field camera techniques with artificial intelligence (AI) technology.
Unlike a conventional camera, the light-field camera contains micro-lens arrays in front of the image sensor, which makes the camera small enough to fit into a smart phone, while allowing it to acquire the spatial and directional information of the light with a single shot. The technique has received attention as it can reconstruct images in a variety of ways including multi-views, refocusing, and 3D image acquisition, giving rise to many potential applications.
However, the optical crosstalk between shadows caused by external light sources in the environment and the micro-lens has limited existing light-field cameras from being able to provide accurate image contrast and 3D reconstruction.
The joint research team applied a vertical-cavity surface-emitting laser (VCSEL) in the near-IR range to stabilize the accuracy of 3D image reconstruction that previously depended on environmental light. When an external light source is shone on a face at 0-, 30-, and 60-degree angles, the light field camera reduces 54% of image reconstruction errors. Additionally, by inserting a light-absorbing layer for visible and near-IR wavelengths between the micro-lens arrays, the team could minimize optical crosstalk while increasing the image contrast by 2.1 times.
Through this technique, the team could overcome the limitations of existing light-field cameras and was able to develop their NIR-based light-field camera (NIR-LFC), optimized for the 3D image reconstruction of facial expressions. Using the NIR-LFC, the team acquired high-quality 3D reconstruction images of facial expressions expressing various emotions regardless of the lighting conditions of the surrounding environment.
The facial expressions in the acquired 3D images were distinguished through machine learning with an average of 85% accuracy – a statistically significant figure compared to when 2D images were used. Furthermore, by calculating the interdependency of distance information that varies with facial expression in 3D images, the team could identify the information a light-field camera utilizes to distinguish human expressions.
Professor Ki-Hun Jeong said, “The sub-miniature light-field camera developed by the research team has the potential to become the new platform to quantitatively analyze the facial expressions and emotions of humans.” To highlight the significance of this research, he added, “It could be applied in various fields including mobile healthcare, field diagnosis, social cognition, and human-machine interactions.”
This research was published in Advanced Intelligent Systems online on December 16, under the title, “Machine-Learned Light-field Camera that Reads Facial Expression from High-Contrast and Illumination Invariant 3D Facial Images.” This research was funded by the Ministry of Science and ICT and the Ministry of Trade, Industry and Energy.
-Publication“Machine-learned light-field camera that reads fascial expression from high-contrast and illumination invariant 3D facial images,” Sang-In Bae, Sangyeon Lee, Jae-Myeong Kwon, Hyun-Kyung Kim. Kyung-Won Jang, Doheon Lee, Ki-Hun Jeong, Advanced Intelligent Systems, December 16, 2021 (doi.org/10.1002/aisy.202100182)
ProfileProfessor Ki-Hun JeongBiophotonic LaboratoryDepartment of Bio and Brain EngineeringKAIST
Professor Doheon LeeDepartment of Bio and Brain EngineeringKAIST
2022.01.21 View 15030 -
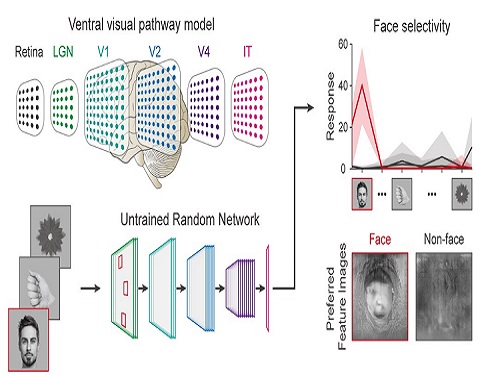 Face Detection in Untrained Deep Neural Networks
A KAIST team shows that primitive visual selectivity of faces can arise spontaneously in completely untrained deep neural networks
Researchers have found that higher visual cognitive functions can arise spontaneously in untrained neural networks. A KAIST research team led by Professor Se-Bum Paik from the Department of Bio and Brain Engineering has shown that visual selectivity of facial images can arise even in completely untrained deep neural networks.
This new finding has provided revelatory insights into mechanisms underlying the development of cognitive functions in both biological and artificial neural networks, also making a significant impact on our understanding of the origin of early brain functions before sensory experiences.
The study published in Nature Communications on December 16 demonstrates that neuronal activities selective to facial images are observed in randomly initialized deep neural networks in the complete absence of learning, and that they show the characteristics of those observed in biological brains.
The ability to identify and recognize faces is a crucial function for social behavior, and this ability is thought to originate from neuronal tuning at the single or multi-neuronal level. Neurons that selectively respond to faces are observed in young animals of various species, and this raises intense debate whether face-selective neurons can arise innately in the brain or if they require visual experience.
Using a model neural network that captures properties of the ventral stream of the visual cortex, the research team found that face-selectivity can emerge spontaneously from random feedforward wirings in untrained deep neural networks. The team showed that the character of this innate face-selectivity is comparable to that observed with face-selective neurons in the brain, and that this spontaneous neuronal tuning for faces enables the network to perform face detection tasks.
These results imply a possible scenario in which the random feedforward connections that develop in early, untrained networks may be sufficient for initializing primitive visual cognitive functions.
Professor Paik said, “Our findings suggest that innate cognitive functions can emerge spontaneously from the statistical complexity embedded in the hierarchical feedforward projection circuitry, even in the complete absence of learning”.
He continued, “Our results provide a broad conceptual advance as well as advanced insight into the mechanisms underlying the development of innate functions in both biological and artificial neural networks, which may unravel the mystery of the generation and evolution of intelligence.” This work was supported by the National Research Foundation of Korea (NRF) and by the KAIST singularity research project.
-PublicationSeungdae Baek, Min Song, Jaeson Jang, Gwangsu Kim, and Se-Bum Baik, “Face detection in untrained deep neural network,” Nature Communications 12, 7328 on Dec.16, 2021
(https://doi.org/10.1038/s41467-021-27606-9)
-ProfileProfessor Se-Bum PaikVisual System and Neural Network LaboratoryProgram of Brain and Cognitive EngineeringDepartment of Bio and Brain EngineeringCollege of EngineeringKAIST
2021.12.21 View 12341
Face Detection in Untrained Deep Neural Networks
A KAIST team shows that primitive visual selectivity of faces can arise spontaneously in completely untrained deep neural networks
Researchers have found that higher visual cognitive functions can arise spontaneously in untrained neural networks. A KAIST research team led by Professor Se-Bum Paik from the Department of Bio and Brain Engineering has shown that visual selectivity of facial images can arise even in completely untrained deep neural networks.
This new finding has provided revelatory insights into mechanisms underlying the development of cognitive functions in both biological and artificial neural networks, also making a significant impact on our understanding of the origin of early brain functions before sensory experiences.
The study published in Nature Communications on December 16 demonstrates that neuronal activities selective to facial images are observed in randomly initialized deep neural networks in the complete absence of learning, and that they show the characteristics of those observed in biological brains.
The ability to identify and recognize faces is a crucial function for social behavior, and this ability is thought to originate from neuronal tuning at the single or multi-neuronal level. Neurons that selectively respond to faces are observed in young animals of various species, and this raises intense debate whether face-selective neurons can arise innately in the brain or if they require visual experience.
Using a model neural network that captures properties of the ventral stream of the visual cortex, the research team found that face-selectivity can emerge spontaneously from random feedforward wirings in untrained deep neural networks. The team showed that the character of this innate face-selectivity is comparable to that observed with face-selective neurons in the brain, and that this spontaneous neuronal tuning for faces enables the network to perform face detection tasks.
These results imply a possible scenario in which the random feedforward connections that develop in early, untrained networks may be sufficient for initializing primitive visual cognitive functions.
Professor Paik said, “Our findings suggest that innate cognitive functions can emerge spontaneously from the statistical complexity embedded in the hierarchical feedforward projection circuitry, even in the complete absence of learning”.
He continued, “Our results provide a broad conceptual advance as well as advanced insight into the mechanisms underlying the development of innate functions in both biological and artificial neural networks, which may unravel the mystery of the generation and evolution of intelligence.” This work was supported by the National Research Foundation of Korea (NRF) and by the KAIST singularity research project.
-PublicationSeungdae Baek, Min Song, Jaeson Jang, Gwangsu Kim, and Se-Bum Baik, “Face detection in untrained deep neural network,” Nature Communications 12, 7328 on Dec.16, 2021
(https://doi.org/10.1038/s41467-021-27606-9)
-ProfileProfessor Se-Bum PaikVisual System and Neural Network LaboratoryProgram of Brain and Cognitive EngineeringDepartment of Bio and Brain EngineeringCollege of EngineeringKAIST
2021.12.21 View 12341 -
 A Study Shows Reactive Electrolyte Additives Improve Lithium Metal Battery Performance
Stable electrode-electrolyte interfaces constructed by fluorine- and nitrogen-donating ionic additives provide an opportunity to improve high-performance lithium metal batteries
A research team showed that electrolyte additives increase the lifetime of lithium metal batteries and remarkably improve the performance of fast charging and discharging. Professor Nam-Soon Choi’s team from the Department of Chemical and Biomolecular Engineering at KAIST hierarchized the solid electrolyte interphase to make a dual-layer structure and showed groundbreaking run times for lithium metal batteries.
The team applied two electrolyte additives that have different reduction and adsorption properties to improve the functionality of the dual-layer solid electrolyte interphase. In addition, the team has confirmed that the structural stability of the nickel-rich cathode was achieved through the formation of a thin protective layer on the cathode. This study was reported in Energy Storage Materials.
Securing high-energy-density lithium metal batteries with a long lifespan and fast charging performance is vital for realizing their ubiquitous use as superior power sources for electric vehicles. Lithium metal batteries comprise a lithium metal anode that delivers 10 times higher capacity than the graphite anodes in lithium-ion batteries. Therefore, lithium metal is an indispensable anode material for realizing high-energy rechargeable batteries. However, undesirable reactions among the electrolytes with lithium metal anodes can reduce the power and this remains an impediment to achieving a longer battery lifespan. Previous studies only focused on the formation of the solid electrolyte interphase on the surface of the lithium metal anode.
The team designed a way to create a dual-layer solid electrolyte interphase to resolve the instability of the lithium metal anode by using electrolyte additives, depending on their electron accepting ability and adsorption tendencies. This hierarchical structure of the solid electrolyte interphase on the lithium metal anode has the potential to be further applied to lithium-alloy anodes, lithium storage structures, and anode-free technology to meet market expectations for electrolyte technology.
The batteries with lithium metal anodes and nickel-rich cathodes represented 80.9% of the initial capacity after 600 cycles and achieved a high Coulombic efficiency of 99.94%. These remarkable results contributed to the development of protective dual-layer solid electrolyte interphase technology for lithium metal anodes.
Professor Choi said that the research suggests a new direction for the development of electrolyte additives to regulate the unstable lithium metal anode-electrolyte interface, the biggest hurdle in research on lithium metal batteries.
She added that anode-free secondary battery technology is expected to be a game changer in the secondary battery market and electrolyte additive technology will contribute to the enhancement of anode-free secondary batteries through the stabilization of lithium metal anodes.
This research was funded by the Technology Development Program to Solve Climate Change of the National Research Foundation in Korea funded by the Ministry of Science, ICT & Future Planning and the Technology Innovation Program funded by the Ministry of Trade, Industry & Energy, and Hyundai Motor Company.
- PublicationSaehun Kim, Sung O Park, Min-Young Lee, Jeong-A Lee, Imanuel Kristanto, Tae Kyung Lee, Daeyeon Hwang, Juyoung Kim, Tae-Ung Wi, Hyun-Wook Lee, Sang Kyu Kwak, and NamSoon Choi, “Stable electrode-electrolyte interfaces constructed by fluorine- and nitrogen-donating ionic additives for high-performance lithium metal batteries,” Energy Storage Materials,45, 1-13 (2022), (doi: https://doi.org/10.1016/j.ensm.2021.10.031)
- ProfileProfessor Nam-Soon ChoiEnergy Materials LaboratoryDepartment of Chemical and Biomolecular EngineeringKAIST
2021.12.16 View 11425
A Study Shows Reactive Electrolyte Additives Improve Lithium Metal Battery Performance
Stable electrode-electrolyte interfaces constructed by fluorine- and nitrogen-donating ionic additives provide an opportunity to improve high-performance lithium metal batteries
A research team showed that electrolyte additives increase the lifetime of lithium metal batteries and remarkably improve the performance of fast charging and discharging. Professor Nam-Soon Choi’s team from the Department of Chemical and Biomolecular Engineering at KAIST hierarchized the solid electrolyte interphase to make a dual-layer structure and showed groundbreaking run times for lithium metal batteries.
The team applied two electrolyte additives that have different reduction and adsorption properties to improve the functionality of the dual-layer solid electrolyte interphase. In addition, the team has confirmed that the structural stability of the nickel-rich cathode was achieved through the formation of a thin protective layer on the cathode. This study was reported in Energy Storage Materials.
Securing high-energy-density lithium metal batteries with a long lifespan and fast charging performance is vital for realizing their ubiquitous use as superior power sources for electric vehicles. Lithium metal batteries comprise a lithium metal anode that delivers 10 times higher capacity than the graphite anodes in lithium-ion batteries. Therefore, lithium metal is an indispensable anode material for realizing high-energy rechargeable batteries. However, undesirable reactions among the electrolytes with lithium metal anodes can reduce the power and this remains an impediment to achieving a longer battery lifespan. Previous studies only focused on the formation of the solid electrolyte interphase on the surface of the lithium metal anode.
The team designed a way to create a dual-layer solid electrolyte interphase to resolve the instability of the lithium metal anode by using electrolyte additives, depending on their electron accepting ability and adsorption tendencies. This hierarchical structure of the solid electrolyte interphase on the lithium metal anode has the potential to be further applied to lithium-alloy anodes, lithium storage structures, and anode-free technology to meet market expectations for electrolyte technology.
The batteries with lithium metal anodes and nickel-rich cathodes represented 80.9% of the initial capacity after 600 cycles and achieved a high Coulombic efficiency of 99.94%. These remarkable results contributed to the development of protective dual-layer solid electrolyte interphase technology for lithium metal anodes.
Professor Choi said that the research suggests a new direction for the development of electrolyte additives to regulate the unstable lithium metal anode-electrolyte interface, the biggest hurdle in research on lithium metal batteries.
She added that anode-free secondary battery technology is expected to be a game changer in the secondary battery market and electrolyte additive technology will contribute to the enhancement of anode-free secondary batteries through the stabilization of lithium metal anodes.
This research was funded by the Technology Development Program to Solve Climate Change of the National Research Foundation in Korea funded by the Ministry of Science, ICT & Future Planning and the Technology Innovation Program funded by the Ministry of Trade, Industry & Energy, and Hyundai Motor Company.
- PublicationSaehun Kim, Sung O Park, Min-Young Lee, Jeong-A Lee, Imanuel Kristanto, Tae Kyung Lee, Daeyeon Hwang, Juyoung Kim, Tae-Ung Wi, Hyun-Wook Lee, Sang Kyu Kwak, and NamSoon Choi, “Stable electrode-electrolyte interfaces constructed by fluorine- and nitrogen-donating ionic additives for high-performance lithium metal batteries,” Energy Storage Materials,45, 1-13 (2022), (doi: https://doi.org/10.1016/j.ensm.2021.10.031)
- ProfileProfessor Nam-Soon ChoiEnergy Materials LaboratoryDepartment of Chemical and Biomolecular EngineeringKAIST
2021.12.16 View 11425 -
 Connecting the Dots to Find New Treatments for Breast Cancer
Systems biologists uncovered new ways of cancer cell reprogramming to treat drug-resistant cancers
Scientists at KAIST believe they may have found a way to reverse an aggressive, treatment-resistant type of breast cancer into a less dangerous kind that responds well to treatment. The study involved the use of mathematical models to untangle the complex genetic and molecular interactions that occur in the two types of breast cancer, but could be extended to find ways for treating many others. The study’s findings were published in the journal Cancer Research.
Basal-like tumours are the most aggressive type of breast cancer, with the worst prognosis. Chemotherapy is the only available treatment option, but patients experience high recurrence rates. On the other hand, luminal-A breast cancer responds well to drugs that specifically target a receptor on their cell surfaces, called estrogen receptor alpha (ERα).
KAIST systems biologist Kwang-Hyun Cho and colleagues analyzed the complex molecular and genetic interactions of basal-like and luminal-A breast cancers to find out if there might be a way to switch the former to the latter and give patients a better chance to respond to treatment.
To do this, they accessed large amounts of cancer and patient data to understand which genes and molecules are involved in the two types. They then input this data into a mathematical model that represents genes, proteins and molecules as dots and the interactions between them as lines. The model can be used to conduct simulations and see how interactions change when certain genes are turned on or off.
“There have been a tremendous number of studies trying to find therapeutic targets for treating basal-like breast cancer patients,” says Cho. “But clinical trials have failed due to the complex and dynamic nature of cancer. To overcome this issue, we looked at breast cancer cells as a complex network system and implemented a systems biological approach to unravel the underlying mechanisms that would allow us to reprogram basal-like into luminal-A breast cancer cells.”
Using this approach, followed by experimental validation on real breast cancer cells, the team found that turning off two key gene regulators, called BCL11A and HDAC1/2, switched a basal-like cancer signalling pathway into a different one used by luminal-A cancer cells. The switch reprograms the cancer cells and makes them more responsive to drugs that target ERα receptors. However, further tests will be needed to confirm that this also works in animal models and eventually humans.
“Our study demonstrates that the systems biological approach can be useful for identifying novel therapeutic targets,” says Cho.
The researchers are now expanding its breast cancer network model to include all breast cancer subtypes. Their ultimate aim is to identify more drug targets and to understand the mechanisms that could drive drug-resistant cells to turn into drug-sensitive ones.
This work was supported by the National Research Foundation of Korea, the Ministry of Science and ICT, Electronics and Telecommunications Research Institute, and the KAIST Grand Challenge 30 Project.
-Publication Sea R. Choi, Chae Young Hwang, Jonghoon Lee, and Kwang-Hyun Cho, “Network Analysis Identifies Regulators of Basal-like Breast Cancer Reprogramming and Endocrine TherapyVulnerability,” Cancer Research, November 30. (doi:10.1158/0008-5472.CAN-21-0621)
-ProfileProfessor Kwang-Hyun ChoLaboratory for Systems Biology and Bio-Inspired EngineeringDepartment of Bio and Brain EngineeringKAIST
2021.12.07 View 13247
Connecting the Dots to Find New Treatments for Breast Cancer
Systems biologists uncovered new ways of cancer cell reprogramming to treat drug-resistant cancers
Scientists at KAIST believe they may have found a way to reverse an aggressive, treatment-resistant type of breast cancer into a less dangerous kind that responds well to treatment. The study involved the use of mathematical models to untangle the complex genetic and molecular interactions that occur in the two types of breast cancer, but could be extended to find ways for treating many others. The study’s findings were published in the journal Cancer Research.
Basal-like tumours are the most aggressive type of breast cancer, with the worst prognosis. Chemotherapy is the only available treatment option, but patients experience high recurrence rates. On the other hand, luminal-A breast cancer responds well to drugs that specifically target a receptor on their cell surfaces, called estrogen receptor alpha (ERα).
KAIST systems biologist Kwang-Hyun Cho and colleagues analyzed the complex molecular and genetic interactions of basal-like and luminal-A breast cancers to find out if there might be a way to switch the former to the latter and give patients a better chance to respond to treatment.
To do this, they accessed large amounts of cancer and patient data to understand which genes and molecules are involved in the two types. They then input this data into a mathematical model that represents genes, proteins and molecules as dots and the interactions between them as lines. The model can be used to conduct simulations and see how interactions change when certain genes are turned on or off.
“There have been a tremendous number of studies trying to find therapeutic targets for treating basal-like breast cancer patients,” says Cho. “But clinical trials have failed due to the complex and dynamic nature of cancer. To overcome this issue, we looked at breast cancer cells as a complex network system and implemented a systems biological approach to unravel the underlying mechanisms that would allow us to reprogram basal-like into luminal-A breast cancer cells.”
Using this approach, followed by experimental validation on real breast cancer cells, the team found that turning off two key gene regulators, called BCL11A and HDAC1/2, switched a basal-like cancer signalling pathway into a different one used by luminal-A cancer cells. The switch reprograms the cancer cells and makes them more responsive to drugs that target ERα receptors. However, further tests will be needed to confirm that this also works in animal models and eventually humans.
“Our study demonstrates that the systems biological approach can be useful for identifying novel therapeutic targets,” says Cho.
The researchers are now expanding its breast cancer network model to include all breast cancer subtypes. Their ultimate aim is to identify more drug targets and to understand the mechanisms that could drive drug-resistant cells to turn into drug-sensitive ones.
This work was supported by the National Research Foundation of Korea, the Ministry of Science and ICT, Electronics and Telecommunications Research Institute, and the KAIST Grand Challenge 30 Project.
-Publication Sea R. Choi, Chae Young Hwang, Jonghoon Lee, and Kwang-Hyun Cho, “Network Analysis Identifies Regulators of Basal-like Breast Cancer Reprogramming and Endocrine TherapyVulnerability,” Cancer Research, November 30. (doi:10.1158/0008-5472.CAN-21-0621)
-ProfileProfessor Kwang-Hyun ChoLaboratory for Systems Biology and Bio-Inspired EngineeringDepartment of Bio and Brain EngineeringKAIST
2021.12.07 View 13247 -
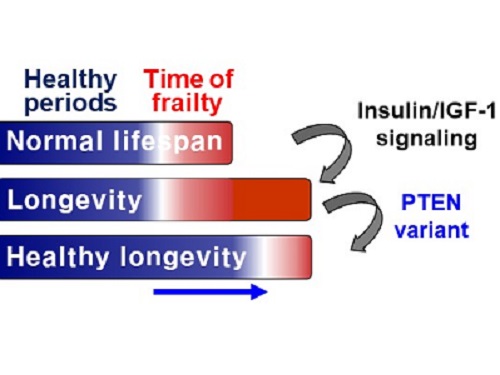 A Genetic Change for Achieving a Long and Healthy Life
Researchers identified a single amino acid change in the tumor suppressor protein in PTEN that extends healthy periods while maintaining longevity
Living a long, healthy life is everyone’s wish, but it is not an easy one to achieve. Many aging studies are developing strategies to increase health spans, the period of life spent with good health, without chronic diseases and disabilities. Researchers at KAIST presented new insights for improving the health span by just regulating the activity of a protein.
A research group under Professor Seung-Jae V. Lee from the Department of Biological Sciences identified a single amino acid change in the tumor suppressor protein phosphatase and tensin homolog (PTEN) that dramatically extends healthy periods while maintaining longevity. This study highlights the importance of the well-conserved tumor suppressor protein PTEN in health span regulation, which can be targeted to develop therapies for promoting healthy longevity in humans. The research was published in Nature Communications on September 24, 2021.
Insulin and insulin-like growth factor-1 (IGF-1) signaling (IIS) is one of the evolutionarily conserved aging-modulatory pathways present in life forms ranging from tiny roundworms to humans. The proper reduction of IIS leads to longevity in animals but often causes defects in multiple health parameters including impaired motility, reproduction, and growth.
The research team found that a specific amino acid change in the PTEN protein improves health status while retaining the longevity conferred by reduced IIS. They used the roundworm C. elegans, an excellent model animal that has been widely used for aging research, mainly because of its very short normal lifespan of about two to three weeks. The PTEN protein is a phosphatase that removes phosphate from lipids as well as proteins. Interestingly, the newly identified amino acid change delicately recalibrated the IIS by partially maintaining protein phosphatase activity while reducing lipid phosphatase activity.
As a result, the amino acid change in the PTEN protein maintained the activity of the longevity-promoting transcription factor Forkhead Box O (FOXO) protein while restricting the detrimental upregulation of another transcription factor, NRF2, leading to long and healthy life in animals with reduced IIS.
Professor Lee said, “Our study raises the exciting possibility of simultaneously promoting longevity and health in humans by slightly tweaking the activity of one protein, PTEN.”
This work was supported by the MInistry of Science and ICT through the National Research Foundation of Korea.
-Publication:Hae-Eun H. Park, Wooseon Hwang, Seokjin Ham, Eunah Kim, Ozlem Altintas, Sangsoon Park, Heehwa G. Son, Yujin Lee, Dongyeop Lee, Won Do Heo, and Seung-Jae V. Lee. 2021. “A PTEN variant uncouples longevity from impaired fitness in Caenorhabditis elegans with reduced insulin/IGF-1 signaling,” Nature Communications, 12(1), 5631. (https://doi.org/10.1038/s41467-021-25920-w)
-ProfileProfessor Seung-Jae V. LeeMolecular Genetics of Aging LaboratoryDepartment of Biological Sciences KAIST
2021.11.19 View 10447
A Genetic Change for Achieving a Long and Healthy Life
Researchers identified a single amino acid change in the tumor suppressor protein in PTEN that extends healthy periods while maintaining longevity
Living a long, healthy life is everyone’s wish, but it is not an easy one to achieve. Many aging studies are developing strategies to increase health spans, the period of life spent with good health, without chronic diseases and disabilities. Researchers at KAIST presented new insights for improving the health span by just regulating the activity of a protein.
A research group under Professor Seung-Jae V. Lee from the Department of Biological Sciences identified a single amino acid change in the tumor suppressor protein phosphatase and tensin homolog (PTEN) that dramatically extends healthy periods while maintaining longevity. This study highlights the importance of the well-conserved tumor suppressor protein PTEN in health span regulation, which can be targeted to develop therapies for promoting healthy longevity in humans. The research was published in Nature Communications on September 24, 2021.
Insulin and insulin-like growth factor-1 (IGF-1) signaling (IIS) is one of the evolutionarily conserved aging-modulatory pathways present in life forms ranging from tiny roundworms to humans. The proper reduction of IIS leads to longevity in animals but often causes defects in multiple health parameters including impaired motility, reproduction, and growth.
The research team found that a specific amino acid change in the PTEN protein improves health status while retaining the longevity conferred by reduced IIS. They used the roundworm C. elegans, an excellent model animal that has been widely used for aging research, mainly because of its very short normal lifespan of about two to three weeks. The PTEN protein is a phosphatase that removes phosphate from lipids as well as proteins. Interestingly, the newly identified amino acid change delicately recalibrated the IIS by partially maintaining protein phosphatase activity while reducing lipid phosphatase activity.
As a result, the amino acid change in the PTEN protein maintained the activity of the longevity-promoting transcription factor Forkhead Box O (FOXO) protein while restricting the detrimental upregulation of another transcription factor, NRF2, leading to long and healthy life in animals with reduced IIS.
Professor Lee said, “Our study raises the exciting possibility of simultaneously promoting longevity and health in humans by slightly tweaking the activity of one protein, PTEN.”
This work was supported by the MInistry of Science and ICT through the National Research Foundation of Korea.
-Publication:Hae-Eun H. Park, Wooseon Hwang, Seokjin Ham, Eunah Kim, Ozlem Altintas, Sangsoon Park, Heehwa G. Son, Yujin Lee, Dongyeop Lee, Won Do Heo, and Seung-Jae V. Lee. 2021. “A PTEN variant uncouples longevity from impaired fitness in Caenorhabditis elegans with reduced insulin/IGF-1 signaling,” Nature Communications, 12(1), 5631. (https://doi.org/10.1038/s41467-021-25920-w)
-ProfileProfessor Seung-Jae V. LeeMolecular Genetics of Aging LaboratoryDepartment of Biological Sciences KAIST
2021.11.19 View 10447 -
 Two Researchers Designated as SUHF Fellows
Professor Taeyun Ku from the Graduate School of Medical Science and Engineering and Professor Hanseul Yang from the Department of Biological Sciences were nominated as 2021 fellows of the Suh Kyungbae Foundation (SUHF).
SUHF selected three young promising scientists from 53 researchers who are less than five years into their careers. A panel of judges comprised of scholars from home and abroad made the final selection based on the candidates’ innovativeness and power to influence. Professor You-Bong Hyun from Seoul National University also won the fellowship.
Professor Ku’s main topic is opto-connectomics. He will study ways to visualize the complex brain network using innovative technology that transforms neurons into optical elements.
Professor Yang will research the possibility of helping patients recover from skin diseases or injuries without scars by studying spiny mouse genes.
SUHF was established by Amorepacific Group Chairman Suh Kyungbae in 2016 with 300 billion KRW of his private funds. Under the vision of ‘contributing to humanity by supporting innovative discoveries of bioscience researchers,’ the foundation supports promising Korean scientists who pioneer new fields of research in biological sciences.
From 2017 to this year, SUHF has selected 20 promising scientists in the field of biological sciences. Selected scientists are provided with up to KRW 500 million each year for five years. The foundation has provided a total of KRW 48.5 billion in research funds to date.
2021.09.15 View 10910
Two Researchers Designated as SUHF Fellows
Professor Taeyun Ku from the Graduate School of Medical Science and Engineering and Professor Hanseul Yang from the Department of Biological Sciences were nominated as 2021 fellows of the Suh Kyungbae Foundation (SUHF).
SUHF selected three young promising scientists from 53 researchers who are less than five years into their careers. A panel of judges comprised of scholars from home and abroad made the final selection based on the candidates’ innovativeness and power to influence. Professor You-Bong Hyun from Seoul National University also won the fellowship.
Professor Ku’s main topic is opto-connectomics. He will study ways to visualize the complex brain network using innovative technology that transforms neurons into optical elements.
Professor Yang will research the possibility of helping patients recover from skin diseases or injuries without scars by studying spiny mouse genes.
SUHF was established by Amorepacific Group Chairman Suh Kyungbae in 2016 with 300 billion KRW of his private funds. Under the vision of ‘contributing to humanity by supporting innovative discoveries of bioscience researchers,’ the foundation supports promising Korean scientists who pioneer new fields of research in biological sciences.
From 2017 to this year, SUHF has selected 20 promising scientists in the field of biological sciences. Selected scientists are provided with up to KRW 500 million each year for five years. The foundation has provided a total of KRW 48.5 billion in research funds to date.
2021.09.15 View 10910 -
 3D Visualization and Quantification of Bioplastic PHA in a Living Bacterial Cell
3D holographic microscopy leads to in-depth analysis of bacterial cells accumulating the bacterial bioplastic, polyhydroxyalkanoate (PHA)
A research team at KAIST has observed how bioplastic granule is being accumulated in living bacteria cells through 3D holographic microscopy. Their 3D imaging and quantitative analysis of the bioplastic ‘polyhydroxyalkanoate’ (PHA) via optical diffraction tomography provides insights into biosynthesizing sustainable substitutes for petroleum-based plastics.
The bio-degradable polyester polyhydroxyalkanoate (PHA) is being touted as an eco-friendly bioplastic to replace existing synthetic plastics. While carrying similar properties to general-purpose plastics such as polyethylene and polypropylene, PHA can be used in various industrial applications such as container packaging and disposable products.
PHA is synthesized by numerous bacteria as an energy and carbon storage material under unbalanced growth conditions in the presence of excess carbon sources. PHA exists in the form of insoluble granules in the cytoplasm. Previous studies on investigating in vivo PHA granules have been performed by using fluorescence microscopy, transmission electron microscopy (TEM), and electron cryotomography.
These techniques have generally relied on the statistical analysis of multiple 2D snapshots of fixed cells or the short-time monitoring of the cells. For the TEM analysis, cells need to be fixed and sectioned, and thus the investigation of living cells was not possible. Fluorescence-based techniques require fluorescence labeling or dye staining. Thus, indirect imaging with the use of reporter proteins cannot show the native state of PHAs or cells, and invasive exogenous dyes can affect the physiology and viability of the cells. Therefore, it was difficult to fully understand the formation of PHA granules in cells due to the technical limitations, and thus several mechanism models based on the observations have been only proposed.
The team of metabolic engineering researchers led by Distinguished Professor Sang Yup Lee and Physics Professor YongKeun Park, who established the startup Tomocube with his 3D holographic microscopy, reported the results of 3D quantitative label-free analysis of PHA granules in individual live bacterial cells by measuring the refractive index distributions using optical diffraction tomography. The formation and growth of PHA granules in the cells of Cupriavidus necator, the most-studied native PHA (specifically, poly(3-hydroxybutyrate), also known as PHB) producer, and recombinant Escherichia coli harboring C. necator PHB biosynthesis pathway were comparatively examined.
From the reconstructed 3D refractive index distribution of the cells, the team succeeded in the 3D visualization and quantitative analysis of cells and intracellular PHA granules at a single-cell level. In particular, the team newly presented the concept of “in vivo PHA granule density.” Through the statistical analysis of hundreds of single cells accumulating PHA granules, the distinctive differences of density and localization of PHA granules in the two micro-organisms were found. Furthermore, the team identified the key protein that plays a major role in making the difference that enabled the characteristics of PHA granules in the recombinant E. coli to become similar to those of C. necator.
The research team also presented 3D time-lapse movies showing the actual processes of PHA granule formation combined with cell growth and division. Movies showing the living cells synthesizing and accumulating PHA granules in their native state had never been reported before.
Professor Lee said, “This study provides insights into the morphological and physical characteristics of in vivo PHA as well as the unique mechanisms of PHA granule formation that undergo the phase transition from soluble monomers into the insoluble polymer, followed by granule formation. Through this study, a deeper understanding of PHA granule formation within the bacterial cells is now possible, which has great significance in that a convergence study of biology and physics was achieved. This study will help develop various bioplastics production processes in the future.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (Grants NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) and the Bio & Medical Technology Development Program (Grant No. 2021M3A9I4022740) from the Ministry of Science and ICT (MSIT) through the National Research Foundation (NRF) of Korea to S.Y.L. This work was also supported by the KAIST Cross-Generation Collaborative Laboratory project.
-PublicationSo Young Choi, Jeonghun Oh, JaeHwang Jung, YongKeun Park, and Sang Yup Lee. Three-dimensional label-free visualization and quantification of polyhydroxyalkanoates in individualbacterial cell in its native state. PNAS(https://doi.org./10.1073/pnas.2103956118)
-ProfileDistinguished Professor Sang Yup LeeMetabolic Engineering and Synthetic Biologyhttp://mbel.kaist.ac.kr/
Department of Chemical and Biomolecular Engineering KAIST
Endowed Chair Professor YongKeun ParkBiomedical Optics Laboratoryhttps://bmokaist.wordpress.com/
Department of PhysicsKAIST
2021.07.28 View 16666
3D Visualization and Quantification of Bioplastic PHA in a Living Bacterial Cell
3D holographic microscopy leads to in-depth analysis of bacterial cells accumulating the bacterial bioplastic, polyhydroxyalkanoate (PHA)
A research team at KAIST has observed how bioplastic granule is being accumulated in living bacteria cells through 3D holographic microscopy. Their 3D imaging and quantitative analysis of the bioplastic ‘polyhydroxyalkanoate’ (PHA) via optical diffraction tomography provides insights into biosynthesizing sustainable substitutes for petroleum-based plastics.
The bio-degradable polyester polyhydroxyalkanoate (PHA) is being touted as an eco-friendly bioplastic to replace existing synthetic plastics. While carrying similar properties to general-purpose plastics such as polyethylene and polypropylene, PHA can be used in various industrial applications such as container packaging and disposable products.
PHA is synthesized by numerous bacteria as an energy and carbon storage material under unbalanced growth conditions in the presence of excess carbon sources. PHA exists in the form of insoluble granules in the cytoplasm. Previous studies on investigating in vivo PHA granules have been performed by using fluorescence microscopy, transmission electron microscopy (TEM), and electron cryotomography.
These techniques have generally relied on the statistical analysis of multiple 2D snapshots of fixed cells or the short-time monitoring of the cells. For the TEM analysis, cells need to be fixed and sectioned, and thus the investigation of living cells was not possible. Fluorescence-based techniques require fluorescence labeling or dye staining. Thus, indirect imaging with the use of reporter proteins cannot show the native state of PHAs or cells, and invasive exogenous dyes can affect the physiology and viability of the cells. Therefore, it was difficult to fully understand the formation of PHA granules in cells due to the technical limitations, and thus several mechanism models based on the observations have been only proposed.
The team of metabolic engineering researchers led by Distinguished Professor Sang Yup Lee and Physics Professor YongKeun Park, who established the startup Tomocube with his 3D holographic microscopy, reported the results of 3D quantitative label-free analysis of PHA granules in individual live bacterial cells by measuring the refractive index distributions using optical diffraction tomography. The formation and growth of PHA granules in the cells of Cupriavidus necator, the most-studied native PHA (specifically, poly(3-hydroxybutyrate), also known as PHB) producer, and recombinant Escherichia coli harboring C. necator PHB biosynthesis pathway were comparatively examined.
From the reconstructed 3D refractive index distribution of the cells, the team succeeded in the 3D visualization and quantitative analysis of cells and intracellular PHA granules at a single-cell level. In particular, the team newly presented the concept of “in vivo PHA granule density.” Through the statistical analysis of hundreds of single cells accumulating PHA granules, the distinctive differences of density and localization of PHA granules in the two micro-organisms were found. Furthermore, the team identified the key protein that plays a major role in making the difference that enabled the characteristics of PHA granules in the recombinant E. coli to become similar to those of C. necator.
The research team also presented 3D time-lapse movies showing the actual processes of PHA granule formation combined with cell growth and division. Movies showing the living cells synthesizing and accumulating PHA granules in their native state had never been reported before.
Professor Lee said, “This study provides insights into the morphological and physical characteristics of in vivo PHA as well as the unique mechanisms of PHA granule formation that undergo the phase transition from soluble monomers into the insoluble polymer, followed by granule formation. Through this study, a deeper understanding of PHA granule formation within the bacterial cells is now possible, which has great significance in that a convergence study of biology and physics was achieved. This study will help develop various bioplastics production processes in the future.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (Grants NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) and the Bio & Medical Technology Development Program (Grant No. 2021M3A9I4022740) from the Ministry of Science and ICT (MSIT) through the National Research Foundation (NRF) of Korea to S.Y.L. This work was also supported by the KAIST Cross-Generation Collaborative Laboratory project.
-PublicationSo Young Choi, Jeonghun Oh, JaeHwang Jung, YongKeun Park, and Sang Yup Lee. Three-dimensional label-free visualization and quantification of polyhydroxyalkanoates in individualbacterial cell in its native state. PNAS(https://doi.org./10.1073/pnas.2103956118)
-ProfileDistinguished Professor Sang Yup LeeMetabolic Engineering and Synthetic Biologyhttp://mbel.kaist.ac.kr/
Department of Chemical and Biomolecular Engineering KAIST
Endowed Chair Professor YongKeun ParkBiomedical Optics Laboratoryhttps://bmokaist.wordpress.com/
Department of PhysicsKAIST
2021.07.28 View 16666