LLO
-
 World's Largest Web Conference To Be Held in Korea
The 2014 International World Wide Web Conference (WWW 2014), the world’s most prestigious academic conference in the field of web, will be held for the first time in Korea. The conference is to be last for five days at Seoul COEX, from 7th to 11th April. International World Wide Web Conference covers a wide range of web-related areas, including technologies, research papers, services and more. Since the first conference in 1994 in Switzerland, it has been held in various parts of North America, Europe, South America and Asia, attracting more than 1000 experts in the field. The 23rd International World Wide Web Conference is managed by the International World Wide Web Conferences Steering Committee (IW3C2) and co-hosted by KAIST and National Agency for Technology and Standards, as well as sponsored by Korea Information Science Society and the World Wide Web Consortium (W3C). Keynote speakers for this year’s conference include inventor of the World Wide Web, Sir Tim Berners-Lee, senior vice president of Microsoft, Dr. Qi Lu, and Carnegie Mellon University’s Prof. Christos Faloutsos, as well as Samsung Electronic’s vice president Jong-Deok Choi. In addition to WWW 2014, BigData Innovators Gathering (BIG 2014) and Web for Access (W4A 2014) is also to be held in joint. KAIST Computer Sciences Department’s Prof. Jinwan Jeong, in charge of directing this year’s conference, said “From one-sided 1st generation web to two-way 2nd generation web, such as blogs, and then recently to the 3rd generation web, which include social networks and semantic webs, the web technologies has grown vastly over the past 25 years. WWW 2014 will be the opportunity for Korea to discuss with the world about the informatization and future of the web.” Pre-registration for WWW 2014 can be applied at the official webpage for WWW 2014 (http://www2014.kr) before 17th February.
2014.02.14 View 11662
World's Largest Web Conference To Be Held in Korea
The 2014 International World Wide Web Conference (WWW 2014), the world’s most prestigious academic conference in the field of web, will be held for the first time in Korea. The conference is to be last for five days at Seoul COEX, from 7th to 11th April. International World Wide Web Conference covers a wide range of web-related areas, including technologies, research papers, services and more. Since the first conference in 1994 in Switzerland, it has been held in various parts of North America, Europe, South America and Asia, attracting more than 1000 experts in the field. The 23rd International World Wide Web Conference is managed by the International World Wide Web Conferences Steering Committee (IW3C2) and co-hosted by KAIST and National Agency for Technology and Standards, as well as sponsored by Korea Information Science Society and the World Wide Web Consortium (W3C). Keynote speakers for this year’s conference include inventor of the World Wide Web, Sir Tim Berners-Lee, senior vice president of Microsoft, Dr. Qi Lu, and Carnegie Mellon University’s Prof. Christos Faloutsos, as well as Samsung Electronic’s vice president Jong-Deok Choi. In addition to WWW 2014, BigData Innovators Gathering (BIG 2014) and Web for Access (W4A 2014) is also to be held in joint. KAIST Computer Sciences Department’s Prof. Jinwan Jeong, in charge of directing this year’s conference, said “From one-sided 1st generation web to two-way 2nd generation web, such as blogs, and then recently to the 3rd generation web, which include social networks and semantic webs, the web technologies has grown vastly over the past 25 years. WWW 2014 will be the opportunity for Korea to discuss with the world about the informatization and future of the web.” Pre-registration for WWW 2014 can be applied at the official webpage for WWW 2014 (http://www2014.kr) before 17th February.
2014.02.14 View 11662 -
 Is it possible to identify rumors on SNS?
Rumors
sporadically spread with people with fewer followers in the centerResearched
over 100 rumors in the US from 2006 to 2009 Is it
possible to filter information on SNS such as Twitter and Facebook? A research
team led by Professor Mee-Young Cha from the Department of Cultural Technology
Graduate School at KAIST, Professor Kyo-Min Jung of Seoul National University,
Doctor Wei Chen and Yajun Wang of Microsoft Asia, has developed a technology
that can accurately filter out information on Twitter to 90% accuracy. The
research not only deduced a new mathematical model, network structure, and
linguistic characteristics on rumors from SNS data, but is also expected to
enhance the effort to make secure technology to regulate Internet rumors. The team
analysed the characteristics of rumors in over 100 widespread cases in the US
from 2006 to 2009 on Twitter. The team gathered data, which included a range of
areas such as politics, IT, health and celebrity gossips, and their analysis
could identify rumors to 90% accuracy. The filtering was more accurate in
rumors that included slanders or insults. The
research team identified three characteristics of the spread of rumors. Firstly,
rumors spread continuously. Normal news spreads widely once and is mentioned
rarely again on media, but rumors tend to continue for years. Secondly,
rumors spread through sporadic participation of random users with no
connections. Rumors start from people with fewer followers and spread to the
more popular. This phenomenon is often observed in rumors concerning
celebrities or politicians. Lastly,
rumors have unique linguistic characteristics. Rumors frequently include words
(such as “it may be true,” “although not certain, I think,” “although I cannot
fully remember”) related to psychological processes that question, deny, or
infer the reliability of the information. Professor
Cha said, “This research deduced not only a statistical and mathematical model
but also is an integrated research on social psychological theory on the
characteristics of rumors that attract great attention from the society based
on ample data.” The results
were made public in IEEE International Conference on Data Mining last December
in Texas, USA.
2014.02.03 View 11347
Is it possible to identify rumors on SNS?
Rumors
sporadically spread with people with fewer followers in the centerResearched
over 100 rumors in the US from 2006 to 2009 Is it
possible to filter information on SNS such as Twitter and Facebook? A research
team led by Professor Mee-Young Cha from the Department of Cultural Technology
Graduate School at KAIST, Professor Kyo-Min Jung of Seoul National University,
Doctor Wei Chen and Yajun Wang of Microsoft Asia, has developed a technology
that can accurately filter out information on Twitter to 90% accuracy. The
research not only deduced a new mathematical model, network structure, and
linguistic characteristics on rumors from SNS data, but is also expected to
enhance the effort to make secure technology to regulate Internet rumors. The team
analysed the characteristics of rumors in over 100 widespread cases in the US
from 2006 to 2009 on Twitter. The team gathered data, which included a range of
areas such as politics, IT, health and celebrity gossips, and their analysis
could identify rumors to 90% accuracy. The filtering was more accurate in
rumors that included slanders or insults. The
research team identified three characteristics of the spread of rumors. Firstly,
rumors spread continuously. Normal news spreads widely once and is mentioned
rarely again on media, but rumors tend to continue for years. Secondly,
rumors spread through sporadic participation of random users with no
connections. Rumors start from people with fewer followers and spread to the
more popular. This phenomenon is often observed in rumors concerning
celebrities or politicians. Lastly,
rumors have unique linguistic characteristics. Rumors frequently include words
(such as “it may be true,” “although not certain, I think,” “although I cannot
fully remember”) related to psychological processes that question, deny, or
infer the reliability of the information. Professor
Cha said, “This research deduced not only a statistical and mathematical model
but also is an integrated research on social psychological theory on the
characteristics of rumors that attract great attention from the society based
on ample data.” The results
were made public in IEEE International Conference on Data Mining last December
in Texas, USA.
2014.02.03 View 11347 -
 A Molecular Switch Controlling Self-Assembly of Protein Nanotubes Discovered
International collaborative research among South Korea, United States, and Israel research institutionsThe key to the treatment of cancer and brain disease mechanism The molecular switch that controls the self-assembly structure of the protein nanotubes, which plays crucial role in cell division and intracellular transport of materials, has been discovered. KAIST Bio and Brain Engineering Department’s Professor Myeong-Cheol Choi and Professor Chae-Yeon Song conducted the research, in collaboration with the University of California in Santa Barbara, U.S., and Hebrew University in Israel. The findings of the research were published in Nature Materials on the 19th. Microtubules are tube shaped and composed of protein that plays a key role in cell division, cytoskeleton, and intercellular material transport and is only 25nm in diameter (1/100,000 thickness of a human hair). Conventionally, cancer treatment focused on disrupting the formation of microtubules to suppress the division of cancer cells. In addition Alzheimer’s is known to be caused by the diminishing of structural integrity of microtubules responsible for intercellular material transport which leads to failure in signal transfer. The research team utilized synchrotron x-ray scattering and transmission electron microscope to analyze the self assemble structure of protein nanotubes to subnanometer accuracy. As a result, the microtubules were found to assemble into 25nm thickness tubules by stacking protein blocks 4 x 5 x 8nm in dimension. In the process, the research team discovered the molecular switch that controls the shape of these protein blocks. In addition the research team was successful in creating a new protein tube structure. Professor Choi commented that they were successful in introducing a new paradigm that suggests the possibility of controlling the complex biological functions of human’s biological system with the simple use of physical principles. He commented further that it is anticipated that the findings will allow for the application of bio nanotubes in engineering and that this is a small step in finding the mechanism behind cancer treatment and neural diseases.
2014.02.03 View 11372
A Molecular Switch Controlling Self-Assembly of Protein Nanotubes Discovered
International collaborative research among South Korea, United States, and Israel research institutionsThe key to the treatment of cancer and brain disease mechanism The molecular switch that controls the self-assembly structure of the protein nanotubes, which plays crucial role in cell division and intracellular transport of materials, has been discovered. KAIST Bio and Brain Engineering Department’s Professor Myeong-Cheol Choi and Professor Chae-Yeon Song conducted the research, in collaboration with the University of California in Santa Barbara, U.S., and Hebrew University in Israel. The findings of the research were published in Nature Materials on the 19th. Microtubules are tube shaped and composed of protein that plays a key role in cell division, cytoskeleton, and intercellular material transport and is only 25nm in diameter (1/100,000 thickness of a human hair). Conventionally, cancer treatment focused on disrupting the formation of microtubules to suppress the division of cancer cells. In addition Alzheimer’s is known to be caused by the diminishing of structural integrity of microtubules responsible for intercellular material transport which leads to failure in signal transfer. The research team utilized synchrotron x-ray scattering and transmission electron microscope to analyze the self assemble structure of protein nanotubes to subnanometer accuracy. As a result, the microtubules were found to assemble into 25nm thickness tubules by stacking protein blocks 4 x 5 x 8nm in dimension. In the process, the research team discovered the molecular switch that controls the shape of these protein blocks. In addition the research team was successful in creating a new protein tube structure. Professor Choi commented that they were successful in introducing a new paradigm that suggests the possibility of controlling the complex biological functions of human’s biological system with the simple use of physical principles. He commented further that it is anticipated that the findings will allow for the application of bio nanotubes in engineering and that this is a small step in finding the mechanism behind cancer treatment and neural diseases.
2014.02.03 View 11372 -
 Professor Suk-Bok Chang receives 14th Korea Science Award in the field of Chemistry
Professor Suk-Bok Chang from the Department of Chemistry at KAIST received the “2013 Korea Science Award” in chemistry hosted by the National Research Foundation and the Ministry of Science, ICT, and Future Planning, Republic of Korea.
The Korea Science Award is a presidential award of Korea, which was first established in 1987 to recognize research excellence in natural science. Three scientists are selected for the award in every other year.
Professor Chang primarily researches the catalyzing mechanism of carbon-hydrogen bonds in organic molecules. He has succeeded in making great progress in the field of organic chemistry especially in developing a new type of transition metal catalytic behavior that can be applied to low-reactivity compounds.
Hydrocarbons are abundant in nature, but its unreactive nature in ambient conditions makes it unsuitable as reactant for compound synthesis. In addition, the mechanism behind transition metal catalyzed carbon-hydrogen bond synthesis has not been proven sufficiently.
The prediction that fossil fuels will be depleted before the end of the century makes hydrocarbon synthesis an extremely important matter.
The need for an effective hydrocarbon synthesis method inspired Professor Chang to pursue research in the transition metal catalysis method and to develop a catalytic system that would allow efficient synthesis even in ambient conditions.
Professor Chang has been the lead researcher for the Institute for Basic Science’s “molecule catalysis reaction research team” since December 2012 and has been carrying out this research in KAIST.
2014.01.27 View 12049
Professor Suk-Bok Chang receives 14th Korea Science Award in the field of Chemistry
Professor Suk-Bok Chang from the Department of Chemistry at KAIST received the “2013 Korea Science Award” in chemistry hosted by the National Research Foundation and the Ministry of Science, ICT, and Future Planning, Republic of Korea.
The Korea Science Award is a presidential award of Korea, which was first established in 1987 to recognize research excellence in natural science. Three scientists are selected for the award in every other year.
Professor Chang primarily researches the catalyzing mechanism of carbon-hydrogen bonds in organic molecules. He has succeeded in making great progress in the field of organic chemistry especially in developing a new type of transition metal catalytic behavior that can be applied to low-reactivity compounds.
Hydrocarbons are abundant in nature, but its unreactive nature in ambient conditions makes it unsuitable as reactant for compound synthesis. In addition, the mechanism behind transition metal catalyzed carbon-hydrogen bond synthesis has not been proven sufficiently.
The prediction that fossil fuels will be depleted before the end of the century makes hydrocarbon synthesis an extremely important matter.
The need for an effective hydrocarbon synthesis method inspired Professor Chang to pursue research in the transition metal catalysis method and to develop a catalytic system that would allow efficient synthesis even in ambient conditions.
Professor Chang has been the lead researcher for the Institute for Basic Science’s “molecule catalysis reaction research team” since December 2012 and has been carrying out this research in KAIST.
2014.01.27 View 12049 -
 Materials Developed for Sodium Rechargeable Battery by EEWS
The research group of Professor William Goddard III, You-Sung Jung, and Jang-Wook Choi from the Graduate School of Energy, Environment, Water, and Sustainability (EEWS) at KAIST has developed a new sodium-ion rechargeable battery which operates at a high voltage, can be charged, and stably discharges over 10,000 cycles. The research results were published in the online version of the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on December 30, 2013.
Since the material costs of sodium rechargeable batteries is 30 to 40 times lower than lithium batteries, it has received attention as an energy saving tool for smart grids and as the next generation of lithium rechargeable batteries. Until now, sodium-ion rechargeable batteries have had issues with stability when charging and discharging. The research group developed a vanadium-based electrode to solve these problems.
The group said follow-up research will be continued to develop advanced technology on sodium rechargeable batteries as it is still currently in the beginning stages.
The research team: From left to right is Professors William Goddard, You-Sung Jung, and Jang-Wook Choi
2014.01.13 View 12136
Materials Developed for Sodium Rechargeable Battery by EEWS
The research group of Professor William Goddard III, You-Sung Jung, and Jang-Wook Choi from the Graduate School of Energy, Environment, Water, and Sustainability (EEWS) at KAIST has developed a new sodium-ion rechargeable battery which operates at a high voltage, can be charged, and stably discharges over 10,000 cycles. The research results were published in the online version of the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on December 30, 2013.
Since the material costs of sodium rechargeable batteries is 30 to 40 times lower than lithium batteries, it has received attention as an energy saving tool for smart grids and as the next generation of lithium rechargeable batteries. Until now, sodium-ion rechargeable batteries have had issues with stability when charging and discharging. The research group developed a vanadium-based electrode to solve these problems.
The group said follow-up research will be continued to develop advanced technology on sodium rechargeable batteries as it is still currently in the beginning stages.
The research team: From left to right is Professors William Goddard, You-Sung Jung, and Jang-Wook Choi
2014.01.13 View 12136 -
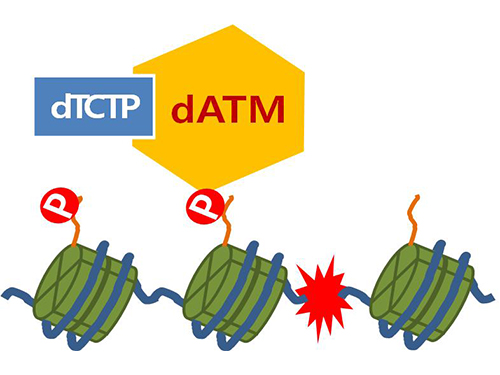 Mechanism in regulation of cancer-related key enzyme, ATM, for DNA damage and repair revealed
Professor Kwang-Wook Choi
A research team led by Professor Kwang-Wook Choi and Dr. Seong-Tae Hong from the Department of Biological Sciences at KAIST has successfully investigated the operational mechanism of the protein Ataxia Telangiectasia Mutated (ATM), an essential protein to the function of a crucial key enzyme that repairs the damaged DNA which stores biometric information. The results were published on December 19th Nature Communications online edition.
All organisms, including humans, constantly strive to protect the information within their DNA from damages posed by a number of factors, such as carbonized materials in our daily food intake, radioactive materials such as radon emitting from the cement of buildings or ultraviolet of the sunlight, which could be a trigger for cancer.
In order to keep the DNA information safe, the organisms are always carrying out complex and sophisticated DNA repair work, which involves the crucial DNA damage repair protein ATM. Consequently, a faulty ATM leads to higher risks of cancer.
Until now, academia predicted that the Translationally Controlled Tumor Protein (TCTP) will play an important role in regulating the function of ATM. However, since most of main research regarding TCTP has only been conducted in cultured cells, it was unable to identify exactly what mechanisms TCTP employs to control ATM.
The KAIST research team identified that TCTP can combine with ATM or increase the enzymatic activity of ATM. In addition, Drosophilia, one of the most widely used model organisms for molecular genetics, has been used to identify that TCTP and ATM play a very important role in repairing the DNA damaged by radiation. This information has allowed the researchers to establish TCTP’s essential function in maintaining the DNA information in cell cultures and even in higher organisms, and to provide specific and important clues to the regulation of ATM by TCTP.
Professor Kwang-Wook Choi said, “Our research is a good example that basic research using Drosophilia can make important contributions to understanding the process of diseases, such as cancer, and to developing adequate treatment.”
The research has been funded by the Ministry of Science, ICT and Future Planning, Republic of Korea, and the National Research Foundation of Korea.
Figure 1. When the amount of TCTP protein is reduced, cells of the Drosophila's eye are abnormally deformed by radiation. Scale bars = 200mm
Figure 2. When the amount of TCTP protein is reduced, the chromosomes of Drosophilia are easily broken by radiation. Scale bars = 10 mm.
Figure 3. When gene expressions of TCTP and ATM are reduced, large defects occur in the normal development of the eye. (Left: normal Drosophilia's eye, right: development-deficient eye)
Figure 4. ATM marks the position of the broken DNA, with TCTP helping to facilitate this reaction. DNA (blue line) within the cell nucleus is coiled around the histone protein (green cylinder). When DNA is broken, ATM protein attaches a phosphate group (P). Multiple DNA repair protein recognizes the phosphate as a signal that requires repair and gathers at the site.
2014.01.07 View 14844
Mechanism in regulation of cancer-related key enzyme, ATM, for DNA damage and repair revealed
Professor Kwang-Wook Choi
A research team led by Professor Kwang-Wook Choi and Dr. Seong-Tae Hong from the Department of Biological Sciences at KAIST has successfully investigated the operational mechanism of the protein Ataxia Telangiectasia Mutated (ATM), an essential protein to the function of a crucial key enzyme that repairs the damaged DNA which stores biometric information. The results were published on December 19th Nature Communications online edition.
All organisms, including humans, constantly strive to protect the information within their DNA from damages posed by a number of factors, such as carbonized materials in our daily food intake, radioactive materials such as radon emitting from the cement of buildings or ultraviolet of the sunlight, which could be a trigger for cancer.
In order to keep the DNA information safe, the organisms are always carrying out complex and sophisticated DNA repair work, which involves the crucial DNA damage repair protein ATM. Consequently, a faulty ATM leads to higher risks of cancer.
Until now, academia predicted that the Translationally Controlled Tumor Protein (TCTP) will play an important role in regulating the function of ATM. However, since most of main research regarding TCTP has only been conducted in cultured cells, it was unable to identify exactly what mechanisms TCTP employs to control ATM.
The KAIST research team identified that TCTP can combine with ATM or increase the enzymatic activity of ATM. In addition, Drosophilia, one of the most widely used model organisms for molecular genetics, has been used to identify that TCTP and ATM play a very important role in repairing the DNA damaged by radiation. This information has allowed the researchers to establish TCTP’s essential function in maintaining the DNA information in cell cultures and even in higher organisms, and to provide specific and important clues to the regulation of ATM by TCTP.
Professor Kwang-Wook Choi said, “Our research is a good example that basic research using Drosophilia can make important contributions to understanding the process of diseases, such as cancer, and to developing adequate treatment.”
The research has been funded by the Ministry of Science, ICT and Future Planning, Republic of Korea, and the National Research Foundation of Korea.
Figure 1. When the amount of TCTP protein is reduced, cells of the Drosophila's eye are abnormally deformed by radiation. Scale bars = 200mm
Figure 2. When the amount of TCTP protein is reduced, the chromosomes of Drosophilia are easily broken by radiation. Scale bars = 10 mm.
Figure 3. When gene expressions of TCTP and ATM are reduced, large defects occur in the normal development of the eye. (Left: normal Drosophilia's eye, right: development-deficient eye)
Figure 4. ATM marks the position of the broken DNA, with TCTP helping to facilitate this reaction. DNA (blue line) within the cell nucleus is coiled around the histone protein (green cylinder). When DNA is broken, ATM protein attaches a phosphate group (P). Multiple DNA repair protein recognizes the phosphate as a signal that requires repair and gathers at the site.
2014.01.07 View 14844 -
 Wearable computer follows suit of smart phones
KAIST hosts “Wearable Computer Competition” in KI Building, Daejeon Campus, on the 7th-8th of November
“Computer that controls smart phones with the movement of facial muscles” and 12 other wearable computers to be presented
As technology transitions to “Wearable Computers,” KAIST is hosting its 9th “Wearable Computer Competition.” The competition will take place over two days, 7th-8th of November, in KI building, on the main Daejeon Campus.
The “wearable computer” is designed to enable users to use the computer whilst moving by limiting its weight and size so that it can be worn as a part of the body and clothing. Wearable computers have been considered the future of information technology (IT) ever since smart phones and other miniaturized IT devices made an appearance.
The “Wearable Computer Competition” has been held since 2005 under the leadership of Professor Hoi-Jun Yoo from the KAIST Department of Electrical Engineering. It is the only competition in the nation where undergraduate students use their unique ideas and newest technology to produce computers that seem to be existed only in sci-fi movies and comic books.
A total of 15 teams out of 70 made the competition and went through a rigorous selection process based on written applications and interviews to enter the final. The teams at the final received USD 1,400 and IT devices including smart phones to produce a wearable computer.
KAIST increased the number of finalists from the last year"s 10 to 15 this year as the wearable computer industry is extending, and there is growing interest in the computer around the world after the launch of Google Glass and Samsung Galaxy Gear.
This year’s entries included a product for quadriplegic patients to control smart phones with the movement of facial muscles, which attracted public interest. The product in the form of a headband can be worn by quadriplegic patients or someone with limited hand movement. The user can activate the product by clenching their molars and move the mouse on the smart phones with the movement of muscles in their face.
Furthermore, a wearable band shaped device that can control smart phones with simple hand movements is also attracting interest. Broad hand movements of the user allows him/her to receive calls and take photos, and handshakes between users control sharing of files. Body communication can be used to protect private information without a password or locking the device.
In addition, gloves and shoes that can sense the user’s movement to play an instrument without the instrument being present; a cane for the blind that converts visual information to tactile; a belt that protects children from sexual crimes; and a game where the user can be Super Mario to play and other practical products are presented.
The chairman of the competition, Professor Yoo said, “As you can see from the launch of Samsung Galaxy Gear, wearable computers will follow smart phones as the leader of IT devices in the next generation.” He continued, “This competition and workshop is an opportunity to increase public interest in wearable computers and serves as a communication platform for experts to view the present and the future of wearable computers.”
The “Wearable Computer Workshop” will be held this year as well. The workshop under the theme of “the present and the future of wearable computers” invited Professor Kyu-Ho Park, Vice President of KAIST, as a keynote speaker to talk on “ubiquitous, fashionable computers.” Moreover, Samsung’s Dong-Jun Geum and the Electronics and Telecommunications Research Institute’s Hyeon-Tae Jeong will lecture on the “trend and direction of progress of wearable devices” and the “technological trend and prospect of industry of wearable computers,” respectively.
To participate in the competition or the workshop, please visit the website (http://www.ufcom.org)
for further information.
2013.11.28 View 11101
Wearable computer follows suit of smart phones
KAIST hosts “Wearable Computer Competition” in KI Building, Daejeon Campus, on the 7th-8th of November
“Computer that controls smart phones with the movement of facial muscles” and 12 other wearable computers to be presented
As technology transitions to “Wearable Computers,” KAIST is hosting its 9th “Wearable Computer Competition.” The competition will take place over two days, 7th-8th of November, in KI building, on the main Daejeon Campus.
The “wearable computer” is designed to enable users to use the computer whilst moving by limiting its weight and size so that it can be worn as a part of the body and clothing. Wearable computers have been considered the future of information technology (IT) ever since smart phones and other miniaturized IT devices made an appearance.
The “Wearable Computer Competition” has been held since 2005 under the leadership of Professor Hoi-Jun Yoo from the KAIST Department of Electrical Engineering. It is the only competition in the nation where undergraduate students use their unique ideas and newest technology to produce computers that seem to be existed only in sci-fi movies and comic books.
A total of 15 teams out of 70 made the competition and went through a rigorous selection process based on written applications and interviews to enter the final. The teams at the final received USD 1,400 and IT devices including smart phones to produce a wearable computer.
KAIST increased the number of finalists from the last year"s 10 to 15 this year as the wearable computer industry is extending, and there is growing interest in the computer around the world after the launch of Google Glass and Samsung Galaxy Gear.
This year’s entries included a product for quadriplegic patients to control smart phones with the movement of facial muscles, which attracted public interest. The product in the form of a headband can be worn by quadriplegic patients or someone with limited hand movement. The user can activate the product by clenching their molars and move the mouse on the smart phones with the movement of muscles in their face.
Furthermore, a wearable band shaped device that can control smart phones with simple hand movements is also attracting interest. Broad hand movements of the user allows him/her to receive calls and take photos, and handshakes between users control sharing of files. Body communication can be used to protect private information without a password or locking the device.
In addition, gloves and shoes that can sense the user’s movement to play an instrument without the instrument being present; a cane for the blind that converts visual information to tactile; a belt that protects children from sexual crimes; and a game where the user can be Super Mario to play and other practical products are presented.
The chairman of the competition, Professor Yoo said, “As you can see from the launch of Samsung Galaxy Gear, wearable computers will follow smart phones as the leader of IT devices in the next generation.” He continued, “This competition and workshop is an opportunity to increase public interest in wearable computers and serves as a communication platform for experts to view the present and the future of wearable computers.”
The “Wearable Computer Workshop” will be held this year as well. The workshop under the theme of “the present and the future of wearable computers” invited Professor Kyu-Ho Park, Vice President of KAIST, as a keynote speaker to talk on “ubiquitous, fashionable computers.” Moreover, Samsung’s Dong-Jun Geum and the Electronics and Telecommunications Research Institute’s Hyeon-Tae Jeong will lecture on the “trend and direction of progress of wearable devices” and the “technological trend and prospect of industry of wearable computers,” respectively.
To participate in the competition or the workshop, please visit the website (http://www.ufcom.org)
for further information.
2013.11.28 View 11101 -
 Cambridge University Press and HISTAC to Publish Science and Civilization in Korea
The KAIST Research Institute for the History of Science, Technology and Civilization of Korea (HISTAC) and Cambridge University Press have agreed to publish a 10-volume collection entitled “Science and Civilization in Korea” in collaboration with the Needham Research Institute.
HISTAC was found in December 2012 with the support of the Academy of Korean Studies and the Korean Studies Promotion Service with the aim of publishing a collection composed of 30 Korean books and 7 English books on Korean science and civilization.
By November 2013, the HISTAC research team submitted a research paper composed of 11 Korean and 1 English book. It has now exceeded its initial goal of publishing 7 English books by signing the recent agreement with the Cambridge University Press.
“Science and Civilization in Korea” is the second collection of non-western science to be published by the Cambridge University Press since 1954 following “Science and Civilization in China” by Joseph Needham who is well-known for his momentous achievements in history of science in East Asia. This collection will highlight the achievements of Korea in science and civilization of Korea, much of which has been under-valued compared to those of China and Japan.[ It now has the significance similar to the Western science and civilization].
HISTAC appointed Professor Hong-Gi Yoon from the University of Auckland as the translator and invited Professor Christopher Cullen from Cambridge University and Professor Morris Low from the University of Queensland as co-editors. Professor Cullen was an editor of “Science and Civilization in China” and is now the director of the Needham Research Institute and Professor Low is an expert in modern science of East Asia.
The series includes:
- History of Science and Technology in Korea
- Technology, Everyday Life, and Korean Civilization
- History and Cultural Studies of Geomancy in Korea
- Patients, Doctors and the State: History of Korean Medical and Pharmaceutical Culture
- History of Astronomy in Korea
- Mathematics and the History of Korean Civilization
- The West and Korea in the History of Science and Technology, 1600-1950
- Imperialism, Colonialism, Post-colonialism and Technological Science in Korea
- Development of Science and Technology Under the Korean Authoritarian Regime
- Dynamics of Technological Development in Korean Industrialization
The HISTAC team believes that the publication will illuminate the nation’s triumphs in science and technology and expects that the publication will serve as valuable research resources for the study of the history of East Asian scientific civilization which has mainly focused on China and Japan. Further, by adopting various case studies of scientific achievements of South Korea and developing countries, they hope to propose a new model for studying history of science and civilization.
2013.11.28 View 9838
Cambridge University Press and HISTAC to Publish Science and Civilization in Korea
The KAIST Research Institute for the History of Science, Technology and Civilization of Korea (HISTAC) and Cambridge University Press have agreed to publish a 10-volume collection entitled “Science and Civilization in Korea” in collaboration with the Needham Research Institute.
HISTAC was found in December 2012 with the support of the Academy of Korean Studies and the Korean Studies Promotion Service with the aim of publishing a collection composed of 30 Korean books and 7 English books on Korean science and civilization.
By November 2013, the HISTAC research team submitted a research paper composed of 11 Korean and 1 English book. It has now exceeded its initial goal of publishing 7 English books by signing the recent agreement with the Cambridge University Press.
“Science and Civilization in Korea” is the second collection of non-western science to be published by the Cambridge University Press since 1954 following “Science and Civilization in China” by Joseph Needham who is well-known for his momentous achievements in history of science in East Asia. This collection will highlight the achievements of Korea in science and civilization of Korea, much of which has been under-valued compared to those of China and Japan.[ It now has the significance similar to the Western science and civilization].
HISTAC appointed Professor Hong-Gi Yoon from the University of Auckland as the translator and invited Professor Christopher Cullen from Cambridge University and Professor Morris Low from the University of Queensland as co-editors. Professor Cullen was an editor of “Science and Civilization in China” and is now the director of the Needham Research Institute and Professor Low is an expert in modern science of East Asia.
The series includes:
- History of Science and Technology in Korea
- Technology, Everyday Life, and Korean Civilization
- History and Cultural Studies of Geomancy in Korea
- Patients, Doctors and the State: History of Korean Medical and Pharmaceutical Culture
- History of Astronomy in Korea
- Mathematics and the History of Korean Civilization
- The West and Korea in the History of Science and Technology, 1600-1950
- Imperialism, Colonialism, Post-colonialism and Technological Science in Korea
- Development of Science and Technology Under the Korean Authoritarian Regime
- Dynamics of Technological Development in Korean Industrialization
The HISTAC team believes that the publication will illuminate the nation’s triumphs in science and technology and expects that the publication will serve as valuable research resources for the study of the history of East Asian scientific civilization which has mainly focused on China and Japan. Further, by adopting various case studies of scientific achievements of South Korea and developing countries, they hope to propose a new model for studying history of science and civilization.
2013.11.28 View 9838 -
 One Day Idea Hackathon, GoGeeks 2013
GoGeeks Creation Group, founded by KAIST alumni, is hosting the GoGeeks One Day Idea Hackathon 2013 on November 23 at the Microsoft (MS) Building in Seoul. The contest was organized to support creative ideas proposed by college students and help them to be applied in real life.
The first GoGeeks contest was held last year by a group of KAIST undergraduate students, which was influenced by Duke University’s Elevator Pitch Competition, an entrepreneurial competition started in 1999.
Applications for the contest should be submitted on www.gogeeks.co.kr by November 20, and the first 30 groups registered will get to compete in the contest.
Each group will make one-minute presentations after six hours of planning on the theme of “Color Your Society.” Then five-minute presentations will follow by the top ten groups selected from the first stage. The top five groups will be each awarded the prize of 2 million Korean won.
The five selected teams will work on their proposed projects during the winter vacation with support from MS Korea and the Center for Science-based Entrepreneurship at KAIST.
Fundraising for the event is going on at http://tumblbug.com/gogeeks2013.
2013.11.21 View 7932
One Day Idea Hackathon, GoGeeks 2013
GoGeeks Creation Group, founded by KAIST alumni, is hosting the GoGeeks One Day Idea Hackathon 2013 on November 23 at the Microsoft (MS) Building in Seoul. The contest was organized to support creative ideas proposed by college students and help them to be applied in real life.
The first GoGeeks contest was held last year by a group of KAIST undergraduate students, which was influenced by Duke University’s Elevator Pitch Competition, an entrepreneurial competition started in 1999.
Applications for the contest should be submitted on www.gogeeks.co.kr by November 20, and the first 30 groups registered will get to compete in the contest.
Each group will make one-minute presentations after six hours of planning on the theme of “Color Your Society.” Then five-minute presentations will follow by the top ten groups selected from the first stage. The top five groups will be each awarded the prize of 2 million Korean won.
The five selected teams will work on their proposed projects during the winter vacation with support from MS Korea and the Center for Science-based Entrepreneurship at KAIST.
Fundraising for the event is going on at http://tumblbug.com/gogeeks2013.
2013.11.21 View 7932 -
 Visit by the President of the University of Illinois at Urbana-Champaign, US
A delegation from the University of Illinois at Urbana-Champaign (UIUC) in the United States visited KAIST on November 11, 2013.
The delegation consisted of senior administrators from the university, the president, vice president for research, dean of engineering college, dean of nursing college, and associate chancellor for corporate and international relations. The managing director of the State of Illinois Far East Office also joined the delegation.
They met with President Steve Kang, including vice presidents and deans of KAIST, and discussed forming a possible partnership between KAIST and UIUC.
Robert A. Easter, the president of UIUC, said, “Higher education institutions today are bearing a critical responsibility to increase the awareness, knowledge, skills, and values needed to create a sustainable future. I hope KAIST and the University of Illinois will join forces to lead innovation in higher education and to stay connected and relevant in a global marketplace.”
President Steve Kang responded, “There would be many opportunities for the two universities to collaborate and achieve global preeminence in such field as biotechnology, engineering, and convergence research.”
The University of Illinois, with three distinct campuses in Chicago, Springfield, and Urbana-Champaign, is one of the most prestigious universities in the world. The university has an annual operating budget of more than USD 5 billion, collectively enrolls more than 78,000 students, awards nearly 20,000 degrees each year, and has more than 665,000 alumni around the world.
2013.11.18 View 8486
Visit by the President of the University of Illinois at Urbana-Champaign, US
A delegation from the University of Illinois at Urbana-Champaign (UIUC) in the United States visited KAIST on November 11, 2013.
The delegation consisted of senior administrators from the university, the president, vice president for research, dean of engineering college, dean of nursing college, and associate chancellor for corporate and international relations. The managing director of the State of Illinois Far East Office also joined the delegation.
They met with President Steve Kang, including vice presidents and deans of KAIST, and discussed forming a possible partnership between KAIST and UIUC.
Robert A. Easter, the president of UIUC, said, “Higher education institutions today are bearing a critical responsibility to increase the awareness, knowledge, skills, and values needed to create a sustainable future. I hope KAIST and the University of Illinois will join forces to lead innovation in higher education and to stay connected and relevant in a global marketplace.”
President Steve Kang responded, “There would be many opportunities for the two universities to collaborate and achieve global preeminence in such field as biotechnology, engineering, and convergence research.”
The University of Illinois, with three distinct campuses in Chicago, Springfield, and Urbana-Champaign, is one of the most prestigious universities in the world. The university has an annual operating budget of more than USD 5 billion, collectively enrolls more than 78,000 students, awards nearly 20,000 degrees each year, and has more than 665,000 alumni around the world.
2013.11.18 View 8486 -
 Professor Ji-Yun Lee, Received FAA Recognition Award
Professor Ji-Yun Lee, from the Department of Aerospace Engineering at KAIST, received the US Federal Aviation Administration (FAA) Recognition Award for her Ground-Based Augmentation System (GBAS) and her contribution to the development of satellite navigation technology.
GBAS contributes to the safety of aircraft navigation by providing flawless information with real-time location accuracy within one meter.
Professor Lee developed the monitoring software to improve the safety of GBAS users in her paper published in the International Journal of Radio Science in July of 2012.
The software will be distributed and used by many organizations including Eurocontrol following verification from the FAA technical center. It is expected to be standardized after discussions among international organizations.Professor Lee said, “As satellite navigation is applied to the infrastructure of air, marine, and ground transportation, as well as information & communications and finance, ensuring the performance and safety of the system is the most important factor. GBAS will be further developed and applied as a part of a global service system through international collaboration.”
2013.11.15 View 12320
Professor Ji-Yun Lee, Received FAA Recognition Award
Professor Ji-Yun Lee, from the Department of Aerospace Engineering at KAIST, received the US Federal Aviation Administration (FAA) Recognition Award for her Ground-Based Augmentation System (GBAS) and her contribution to the development of satellite navigation technology.
GBAS contributes to the safety of aircraft navigation by providing flawless information with real-time location accuracy within one meter.
Professor Lee developed the monitoring software to improve the safety of GBAS users in her paper published in the International Journal of Radio Science in July of 2012.
The software will be distributed and used by many organizations including Eurocontrol following verification from the FAA technical center. It is expected to be standardized after discussions among international organizations.Professor Lee said, “As satellite navigation is applied to the infrastructure of air, marine, and ground transportation, as well as information & communications and finance, ensuring the performance and safety of the system is the most important factor. GBAS will be further developed and applied as a part of a global service system through international collaboration.”
2013.11.15 View 12320 -
 2013 International Forum on Eco-Friendly Vehicle and System
Leaders in transportation technology gathered at KAIST to discuss commercialization & standardization and to encourage the exchange of research progress, strategy, and future initiatives in transportation technology.
The Graduate School for Green Transportation at KAIST hosted the 2013 International Forum on Eco-friendly Vehicles and Systems (IFEV) in Fusion Hall of the KAIST Institute Building from October 21 to 22.
About 50 leaders in the field of future transportation from academic institutes and industries including Dr. Soon-Man Hong, President of Korea Railroad Research Institute (KRRI), Dr. Kwang-Hee Nam, Professor at Pohang University of Science and Technology (POSTECH), and Mr. Mike Schagrin, the Intelligent Transportation Systems Program Manager of the US Department of Transportation (retired) participated in the 4th annual IFEV.
The commercialization & standardization session and a technical session were followed by the plenary meeting of the forum.
Dr. Hong, the keynote speaker, introduced the High Capacity Double Deck High Speed Train, Near Surface Subway System, and Urban Railway System with Wireless Power Transfer Technology under the title “Korea’s Policy and Technology Initiative for Enhancing Green Transport Systems.”
Dr. Kwang-Hee Nam presented “Electric Vehicle Trends & the POSTECH E-Car Research Center Power Train Design,” followed by Mr. Mike Schagrin who spoke about “Going Green with Connected Automation.”
Dr. Omer C. Onar from the Oak Ridge National Laboratory (ORNL) shared recent research on “ORNL Development in Stationary and Dynamic Wireless Charging.”
In the commercialization session, Faical Turki of Vahle, Germany, presented “Wireless Inductive Battery Chargers,” and Professor Kazuyuki Ouchi from Tokyo University presented “Wind Challenger, the Next Generation Hybrid Vessels.”
In the technical session, presentations and discussions were performed on future ground vehicles and railroad technology, intelligent transportation systems and strategy, and policy on eco-friendly vehicle technology, including Professor In-Soo Suh of the Graduate School for Green Transportation at KAIST who presented on “Armadillo-T: 4WD Micro Electric EV with a Foldable Body Concept.”
On the second day of IFEV 2013, representatives of the European Union’s Safe and Green Road Vehicles (SAGE) consortium discussed connectivity in road transportation as a means of improving safety, efficiency and convenience in future safe and green vehicles with collaboration from Korean transportation organizations such as the Korea Transport Institute and Electronics and Telecommunications Research Institute.
Professor Suh, who organized the forum, said, “This forum will serve as an excellent opportunity to discuss and share R&BD progress in the green transportation field. “Details can be found at http://gt.kaist.ac.kr/ifev2013/.
2013.11.15 View 13677
2013 International Forum on Eco-Friendly Vehicle and System
Leaders in transportation technology gathered at KAIST to discuss commercialization & standardization and to encourage the exchange of research progress, strategy, and future initiatives in transportation technology.
The Graduate School for Green Transportation at KAIST hosted the 2013 International Forum on Eco-friendly Vehicles and Systems (IFEV) in Fusion Hall of the KAIST Institute Building from October 21 to 22.
About 50 leaders in the field of future transportation from academic institutes and industries including Dr. Soon-Man Hong, President of Korea Railroad Research Institute (KRRI), Dr. Kwang-Hee Nam, Professor at Pohang University of Science and Technology (POSTECH), and Mr. Mike Schagrin, the Intelligent Transportation Systems Program Manager of the US Department of Transportation (retired) participated in the 4th annual IFEV.
The commercialization & standardization session and a technical session were followed by the plenary meeting of the forum.
Dr. Hong, the keynote speaker, introduced the High Capacity Double Deck High Speed Train, Near Surface Subway System, and Urban Railway System with Wireless Power Transfer Technology under the title “Korea’s Policy and Technology Initiative for Enhancing Green Transport Systems.”
Dr. Kwang-Hee Nam presented “Electric Vehicle Trends & the POSTECH E-Car Research Center Power Train Design,” followed by Mr. Mike Schagrin who spoke about “Going Green with Connected Automation.”
Dr. Omer C. Onar from the Oak Ridge National Laboratory (ORNL) shared recent research on “ORNL Development in Stationary and Dynamic Wireless Charging.”
In the commercialization session, Faical Turki of Vahle, Germany, presented “Wireless Inductive Battery Chargers,” and Professor Kazuyuki Ouchi from Tokyo University presented “Wind Challenger, the Next Generation Hybrid Vessels.”
In the technical session, presentations and discussions were performed on future ground vehicles and railroad technology, intelligent transportation systems and strategy, and policy on eco-friendly vehicle technology, including Professor In-Soo Suh of the Graduate School for Green Transportation at KAIST who presented on “Armadillo-T: 4WD Micro Electric EV with a Foldable Body Concept.”
On the second day of IFEV 2013, representatives of the European Union’s Safe and Green Road Vehicles (SAGE) consortium discussed connectivity in road transportation as a means of improving safety, efficiency and convenience in future safe and green vehicles with collaboration from Korean transportation organizations such as the Korea Transport Institute and Electronics and Telecommunications Research Institute.
Professor Suh, who organized the forum, said, “This forum will serve as an excellent opportunity to discuss and share R&BD progress in the green transportation field. “Details can be found at http://gt.kaist.ac.kr/ifev2013/.
2013.11.15 View 13677