Sang+Yup+Lee
-
 VP Sang Yup Lee Receives Honorary Doctorate from DTU
Vice President for Research, Distinguished Professor Sang Yup Lee at the Department of Chemical & Biomolecular Engineering, was awarded an honorary doctorate from the Technical University of Denmark (DTU) during the DTU Commemoration Day 2022 on April 29. The event drew distinguished guests, students, and faculty including HRH The Crown Prince Frederik Andre Henrik Christian and DTU President Anders Bjarklev.
Professor Lee was recognized for his exceptional scholarship in the field of systems metabolic engineering, which led to the development of microcell factories capable of producing a wide range of fuels, chemicals, materials, and natural compounds, many for the first time.
Professor Lee said in his acceptance speech that KAIST’s continued partnership with DTU in the field of biotechnology will lead to significant contributions in the global efforts to respond to climate change and promote green growth.
DTU CPO and CSO Dina Petronovic Nielson, who heads DTU Biosustain, also lauded Professor Lee saying, “It is not only a great honor for Professor Lee to be induced at DTU but also great honor for DTU to have him.”
Professor Lee also gave commemorative lectures at DTU Biosustain in Lingby and the Bio Innovation Research Institute at the Novo Nordisk Foundation in Copenhagen while in Denmark.
DTU, one of the leading science and technology universities in Europe, has been awarding honorary doctorates since 1921, including to Nobel laureate in chemistry Professor Frances Arnold at Caltech. Professor Lee is the first Korean to receive an honorary doctorate from DTU.
2022.05.03 View 13302
VP Sang Yup Lee Receives Honorary Doctorate from DTU
Vice President for Research, Distinguished Professor Sang Yup Lee at the Department of Chemical & Biomolecular Engineering, was awarded an honorary doctorate from the Technical University of Denmark (DTU) during the DTU Commemoration Day 2022 on April 29. The event drew distinguished guests, students, and faculty including HRH The Crown Prince Frederik Andre Henrik Christian and DTU President Anders Bjarklev.
Professor Lee was recognized for his exceptional scholarship in the field of systems metabolic engineering, which led to the development of microcell factories capable of producing a wide range of fuels, chemicals, materials, and natural compounds, many for the first time.
Professor Lee said in his acceptance speech that KAIST’s continued partnership with DTU in the field of biotechnology will lead to significant contributions in the global efforts to respond to climate change and promote green growth.
DTU CPO and CSO Dina Petronovic Nielson, who heads DTU Biosustain, also lauded Professor Lee saying, “It is not only a great honor for Professor Lee to be induced at DTU but also great honor for DTU to have him.”
Professor Lee also gave commemorative lectures at DTU Biosustain in Lingby and the Bio Innovation Research Institute at the Novo Nordisk Foundation in Copenhagen while in Denmark.
DTU, one of the leading science and technology universities in Europe, has been awarding honorary doctorates since 1921, including to Nobel laureate in chemistry Professor Frances Arnold at Caltech. Professor Lee is the first Korean to receive an honorary doctorate from DTU.
2022.05.03 View 13302 -
 Five Projects Ranked in the Top 100 for National R&D Excellence
Five KAIST research projects were selected as the 2021 Top 100 for National R&D Excellence by the Ministry of Science and ICT and the Korea Institute of Science & Technology Evaluation and Planning.
The five projects are:-The development of E. coli that proliferates with only formic acid and carbon dioxide by Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering
-An original reverse aging technology that restores an old human skin cell into a younger one by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering-The development of next-generation high-efficiency perovskite-silicon tandem solar cells by Professor Byungha Shin from the Department of Materials Science and Engineering-Research on the effects of ultrafine dust in the atmosphere has on energy consumption by Professor Jiyong Eom from the School of Business and Technology Management-Research on a molecular trigger that controls the phase transformation of bio materials by Professor Myungchul Kim from the Department of Bio and Brain Engineering
Started in 2006, an Evaluation Committee composed of experts in industries, universities, and research institutes has made the preliminary selections of the most outstanding research projects based on their significance as a scientific and technological development and their socioeconomic effects. The finalists went through an open public evaluation. The final 100 studies are from six fields: 18 from mechanics & materials, 26 from biology & marine sciences, 19 from ICT & electronics, 10 from interdisciplinary research, and nine from natural science and infrastructure.
The selected 100 studies will receive a certificate and an award plaque from the minister of MSIT as well as additional points for business and institutional evaluations according to appropriate regulations, and the selected researchers will be strongly recommended as candidates for national meritorious awards.
In particular, to help the 100 selected research projects become more accessible for the general public, their main contents will be provided in a free e-book ‘The Top 100 for National R&D Excellence of 2021’ that will be available from online booksellers.
2022.02.17 View 12949
Five Projects Ranked in the Top 100 for National R&D Excellence
Five KAIST research projects were selected as the 2021 Top 100 for National R&D Excellence by the Ministry of Science and ICT and the Korea Institute of Science & Technology Evaluation and Planning.
The five projects are:-The development of E. coli that proliferates with only formic acid and carbon dioxide by Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering
-An original reverse aging technology that restores an old human skin cell into a younger one by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering-The development of next-generation high-efficiency perovskite-silicon tandem solar cells by Professor Byungha Shin from the Department of Materials Science and Engineering-Research on the effects of ultrafine dust in the atmosphere has on energy consumption by Professor Jiyong Eom from the School of Business and Technology Management-Research on a molecular trigger that controls the phase transformation of bio materials by Professor Myungchul Kim from the Department of Bio and Brain Engineering
Started in 2006, an Evaluation Committee composed of experts in industries, universities, and research institutes has made the preliminary selections of the most outstanding research projects based on their significance as a scientific and technological development and their socioeconomic effects. The finalists went through an open public evaluation. The final 100 studies are from six fields: 18 from mechanics & materials, 26 from biology & marine sciences, 19 from ICT & electronics, 10 from interdisciplinary research, and nine from natural science and infrastructure.
The selected 100 studies will receive a certificate and an award plaque from the minister of MSIT as well as additional points for business and institutional evaluations according to appropriate regulations, and the selected researchers will be strongly recommended as candidates for national meritorious awards.
In particular, to help the 100 selected research projects become more accessible for the general public, their main contents will be provided in a free e-book ‘The Top 100 for National R&D Excellence of 2021’ that will be available from online booksellers.
2022.02.17 View 12949 -
 3D Visualization and Quantification of Bioplastic PHA in a Living Bacterial Cell
3D holographic microscopy leads to in-depth analysis of bacterial cells accumulating the bacterial bioplastic, polyhydroxyalkanoate (PHA)
A research team at KAIST has observed how bioplastic granule is being accumulated in living bacteria cells through 3D holographic microscopy. Their 3D imaging and quantitative analysis of the bioplastic ‘polyhydroxyalkanoate’ (PHA) via optical diffraction tomography provides insights into biosynthesizing sustainable substitutes for petroleum-based plastics.
The bio-degradable polyester polyhydroxyalkanoate (PHA) is being touted as an eco-friendly bioplastic to replace existing synthetic plastics. While carrying similar properties to general-purpose plastics such as polyethylene and polypropylene, PHA can be used in various industrial applications such as container packaging and disposable products.
PHA is synthesized by numerous bacteria as an energy and carbon storage material under unbalanced growth conditions in the presence of excess carbon sources. PHA exists in the form of insoluble granules in the cytoplasm. Previous studies on investigating in vivo PHA granules have been performed by using fluorescence microscopy, transmission electron microscopy (TEM), and electron cryotomography.
These techniques have generally relied on the statistical analysis of multiple 2D snapshots of fixed cells or the short-time monitoring of the cells. For the TEM analysis, cells need to be fixed and sectioned, and thus the investigation of living cells was not possible. Fluorescence-based techniques require fluorescence labeling or dye staining. Thus, indirect imaging with the use of reporter proteins cannot show the native state of PHAs or cells, and invasive exogenous dyes can affect the physiology and viability of the cells. Therefore, it was difficult to fully understand the formation of PHA granules in cells due to the technical limitations, and thus several mechanism models based on the observations have been only proposed.
The team of metabolic engineering researchers led by Distinguished Professor Sang Yup Lee and Physics Professor YongKeun Park, who established the startup Tomocube with his 3D holographic microscopy, reported the results of 3D quantitative label-free analysis of PHA granules in individual live bacterial cells by measuring the refractive index distributions using optical diffraction tomography. The formation and growth of PHA granules in the cells of Cupriavidus necator, the most-studied native PHA (specifically, poly(3-hydroxybutyrate), also known as PHB) producer, and recombinant Escherichia coli harboring C. necator PHB biosynthesis pathway were comparatively examined.
From the reconstructed 3D refractive index distribution of the cells, the team succeeded in the 3D visualization and quantitative analysis of cells and intracellular PHA granules at a single-cell level. In particular, the team newly presented the concept of “in vivo PHA granule density.” Through the statistical analysis of hundreds of single cells accumulating PHA granules, the distinctive differences of density and localization of PHA granules in the two micro-organisms were found. Furthermore, the team identified the key protein that plays a major role in making the difference that enabled the characteristics of PHA granules in the recombinant E. coli to become similar to those of C. necator.
The research team also presented 3D time-lapse movies showing the actual processes of PHA granule formation combined with cell growth and division. Movies showing the living cells synthesizing and accumulating PHA granules in their native state had never been reported before.
Professor Lee said, “This study provides insights into the morphological and physical characteristics of in vivo PHA as well as the unique mechanisms of PHA granule formation that undergo the phase transition from soluble monomers into the insoluble polymer, followed by granule formation. Through this study, a deeper understanding of PHA granule formation within the bacterial cells is now possible, which has great significance in that a convergence study of biology and physics was achieved. This study will help develop various bioplastics production processes in the future.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (Grants NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) and the Bio & Medical Technology Development Program (Grant No. 2021M3A9I4022740) from the Ministry of Science and ICT (MSIT) through the National Research Foundation (NRF) of Korea to S.Y.L. This work was also supported by the KAIST Cross-Generation Collaborative Laboratory project.
-PublicationSo Young Choi, Jeonghun Oh, JaeHwang Jung, YongKeun Park, and Sang Yup Lee. Three-dimensional label-free visualization and quantification of polyhydroxyalkanoates in individualbacterial cell in its native state. PNAS(https://doi.org./10.1073/pnas.2103956118)
-ProfileDistinguished Professor Sang Yup LeeMetabolic Engineering and Synthetic Biologyhttp://mbel.kaist.ac.kr/
Department of Chemical and Biomolecular Engineering KAIST
Endowed Chair Professor YongKeun ParkBiomedical Optics Laboratoryhttps://bmokaist.wordpress.com/
Department of PhysicsKAIST
2021.07.28 View 16628
3D Visualization and Quantification of Bioplastic PHA in a Living Bacterial Cell
3D holographic microscopy leads to in-depth analysis of bacterial cells accumulating the bacterial bioplastic, polyhydroxyalkanoate (PHA)
A research team at KAIST has observed how bioplastic granule is being accumulated in living bacteria cells through 3D holographic microscopy. Their 3D imaging and quantitative analysis of the bioplastic ‘polyhydroxyalkanoate’ (PHA) via optical diffraction tomography provides insights into biosynthesizing sustainable substitutes for petroleum-based plastics.
The bio-degradable polyester polyhydroxyalkanoate (PHA) is being touted as an eco-friendly bioplastic to replace existing synthetic plastics. While carrying similar properties to general-purpose plastics such as polyethylene and polypropylene, PHA can be used in various industrial applications such as container packaging and disposable products.
PHA is synthesized by numerous bacteria as an energy and carbon storage material under unbalanced growth conditions in the presence of excess carbon sources. PHA exists in the form of insoluble granules in the cytoplasm. Previous studies on investigating in vivo PHA granules have been performed by using fluorescence microscopy, transmission electron microscopy (TEM), and electron cryotomography.
These techniques have generally relied on the statistical analysis of multiple 2D snapshots of fixed cells or the short-time monitoring of the cells. For the TEM analysis, cells need to be fixed and sectioned, and thus the investigation of living cells was not possible. Fluorescence-based techniques require fluorescence labeling or dye staining. Thus, indirect imaging with the use of reporter proteins cannot show the native state of PHAs or cells, and invasive exogenous dyes can affect the physiology and viability of the cells. Therefore, it was difficult to fully understand the formation of PHA granules in cells due to the technical limitations, and thus several mechanism models based on the observations have been only proposed.
The team of metabolic engineering researchers led by Distinguished Professor Sang Yup Lee and Physics Professor YongKeun Park, who established the startup Tomocube with his 3D holographic microscopy, reported the results of 3D quantitative label-free analysis of PHA granules in individual live bacterial cells by measuring the refractive index distributions using optical diffraction tomography. The formation and growth of PHA granules in the cells of Cupriavidus necator, the most-studied native PHA (specifically, poly(3-hydroxybutyrate), also known as PHB) producer, and recombinant Escherichia coli harboring C. necator PHB biosynthesis pathway were comparatively examined.
From the reconstructed 3D refractive index distribution of the cells, the team succeeded in the 3D visualization and quantitative analysis of cells and intracellular PHA granules at a single-cell level. In particular, the team newly presented the concept of “in vivo PHA granule density.” Through the statistical analysis of hundreds of single cells accumulating PHA granules, the distinctive differences of density and localization of PHA granules in the two micro-organisms were found. Furthermore, the team identified the key protein that plays a major role in making the difference that enabled the characteristics of PHA granules in the recombinant E. coli to become similar to those of C. necator.
The research team also presented 3D time-lapse movies showing the actual processes of PHA granule formation combined with cell growth and division. Movies showing the living cells synthesizing and accumulating PHA granules in their native state had never been reported before.
Professor Lee said, “This study provides insights into the morphological and physical characteristics of in vivo PHA as well as the unique mechanisms of PHA granule formation that undergo the phase transition from soluble monomers into the insoluble polymer, followed by granule formation. Through this study, a deeper understanding of PHA granule formation within the bacterial cells is now possible, which has great significance in that a convergence study of biology and physics was achieved. This study will help develop various bioplastics production processes in the future.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (Grants NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) and the Bio & Medical Technology Development Program (Grant No. 2021M3A9I4022740) from the Ministry of Science and ICT (MSIT) through the National Research Foundation (NRF) of Korea to S.Y.L. This work was also supported by the KAIST Cross-Generation Collaborative Laboratory project.
-PublicationSo Young Choi, Jeonghun Oh, JaeHwang Jung, YongKeun Park, and Sang Yup Lee. Three-dimensional label-free visualization and quantification of polyhydroxyalkanoates in individualbacterial cell in its native state. PNAS(https://doi.org./10.1073/pnas.2103956118)
-ProfileDistinguished Professor Sang Yup LeeMetabolic Engineering and Synthetic Biologyhttp://mbel.kaist.ac.kr/
Department of Chemical and Biomolecular Engineering KAIST
Endowed Chair Professor YongKeun ParkBiomedical Optics Laboratoryhttps://bmokaist.wordpress.com/
Department of PhysicsKAIST
2021.07.28 View 16628 -
 VP Sang Yup Lee Honored with the Pony Chung Innovation Award
Vice President for Research Sang Yup Lee became the recipient of the Innovation Award by the Pony Chung Foundation that was established to honor the late Se-yung Chung, the former chairman of Hyundai Development Company. He will receive 200 million KRW in prize money. Chairman Chung developed Korea’s first domestically manufactured automobile, ‘Pony,’ in the mid-1970s that became the cornerstone of Korea’s auto industry today.
Distinguished Professor Lee, from the Department of Chemical and Biomolecular Engineering, is a pioneering scholar in the field of systems metabolic engineering who developed various micro-organisms for producing a wide range of fuels, chemicals, materials, and natural compounds.
He recently was elected as a foreign member of the Royal Society in the UK and is the first Korean ever elected into the National Academy of Inventors (NAI) in the US as well as one of 13 scholars elected as an International Member of both the National Academy of Sciences (NAS) and the National Academy of Engineering (NAE) in the US.
2021.07.13 View 13010
VP Sang Yup Lee Honored with the Pony Chung Innovation Award
Vice President for Research Sang Yup Lee became the recipient of the Innovation Award by the Pony Chung Foundation that was established to honor the late Se-yung Chung, the former chairman of Hyundai Development Company. He will receive 200 million KRW in prize money. Chairman Chung developed Korea’s first domestically manufactured automobile, ‘Pony,’ in the mid-1970s that became the cornerstone of Korea’s auto industry today.
Distinguished Professor Lee, from the Department of Chemical and Biomolecular Engineering, is a pioneering scholar in the field of systems metabolic engineering who developed various micro-organisms for producing a wide range of fuels, chemicals, materials, and natural compounds.
He recently was elected as a foreign member of the Royal Society in the UK and is the first Korean ever elected into the National Academy of Inventors (NAI) in the US as well as one of 13 scholars elected as an International Member of both the National Academy of Sciences (NAS) and the National Academy of Engineering (NAE) in the US.
2021.07.13 View 13010 -
 Repurposed Drugs Present New Strategy for Treating COVID-19
Virtual screening of 6,218 drugs and cell-based assays identifies best therapeutic medication candidates
A joint research group from KAIST and Institut Pasteur Korea has identified repurposed drugs for COVID-19 treatment through virtual screening and cell-based assays. The research team suggested the strategy for virtual screening with greatly reduced false positives by incorporating pre-docking filtering based on shape similarity and post-docking filtering based on interaction similarity. This strategy will help develop therapeutic medications for COVID-19 and other antiviral diseases more rapidly. This study was reported at the Proceedings of the National Academy of Sciences of the United States of America (PNAS).
Researchers screened 6,218 drugs from a collection of FDA-approved drugs or those under clinical trial and identified 38 potential repurposed drugs for COVID-19 with this strategy. Among them, seven compounds inhibited SARS-CoV-2 replication in Vero cells. Three of these drugs, emodin, omipalisib, and tipifarnib, showed anti-SARS-CoV-2 activity in human lung cells, Calu-3.
Drug repurposing is a practical strategy for developing antiviral drugs in a short period of time, especially during a global pandemic. In many instances, drug repurposing starts with the virtual screening of approved drugs. However, the actual hit rate of virtual screening is low and most of the predicted drug candidates are false positives.
The research group developed effective filtering algorithms before and after the docking simulations to improve the hit rates. In the pre-docking filtering process, compounds with similar shapes to the known active compounds for each target protein were selected and used for docking simulations. In the post-docking filtering process, the chemicals identified through their docking simulations were evaluated considering the docking energy and the similarity of the protein-ligand interactions with the known active compounds.
The experimental results showed that the virtual screening strategy reached a high hit rate of 18.4%, leading to the identification of seven potential drugs out of the 38 drugs initially selected.
“We plan to conduct further preclinical trials for optimizing drug concentrations as one of the three candidates didn’t resolve the toxicity issues in preclinical trials,” said Woo Dae Jang, one of the researchers from KAIST.
“The most important part of this research is that we developed a platform technology that can rapidly identify novel compounds for COVID-19 treatment. If we use this technology, we will be able to quickly respond to new infectious diseases as well as variants of the coronavirus,” said Distinguished Professor Sang Yup Lee.
This work was supported by the KAIST Mobile Clinic Module Project funded by the Ministry of Science and ICT (MSIT) and the National Research Foundation of Korea (NRF). The National Culture Collection for Pathogens in Korea provided the SARS-CoV-2 (NCCP43326).
-PublicationWoo Dae Jang, Sangeun Jeon, Seungtaek Kim, and Sang Yup Lee. Drugs repurposed for COVID-19 by virtual screening of 6,218 drugs and cell-based assay. Proc. Natl. Acad. Sci. U.S.A. (https://doi/org/10.1073/pnas.2024302118)
-ProfileDistinguished Professor Sang Yup LeeMetabolic &Biomolecular Engineering National Research Laboratoryhttp://mbel.kaist.ac.kr
Department of Chemical and Biomolecular EngineeringKAIST
2021.07.08 View 17292
Repurposed Drugs Present New Strategy for Treating COVID-19
Virtual screening of 6,218 drugs and cell-based assays identifies best therapeutic medication candidates
A joint research group from KAIST and Institut Pasteur Korea has identified repurposed drugs for COVID-19 treatment through virtual screening and cell-based assays. The research team suggested the strategy for virtual screening with greatly reduced false positives by incorporating pre-docking filtering based on shape similarity and post-docking filtering based on interaction similarity. This strategy will help develop therapeutic medications for COVID-19 and other antiviral diseases more rapidly. This study was reported at the Proceedings of the National Academy of Sciences of the United States of America (PNAS).
Researchers screened 6,218 drugs from a collection of FDA-approved drugs or those under clinical trial and identified 38 potential repurposed drugs for COVID-19 with this strategy. Among them, seven compounds inhibited SARS-CoV-2 replication in Vero cells. Three of these drugs, emodin, omipalisib, and tipifarnib, showed anti-SARS-CoV-2 activity in human lung cells, Calu-3.
Drug repurposing is a practical strategy for developing antiviral drugs in a short period of time, especially during a global pandemic. In many instances, drug repurposing starts with the virtual screening of approved drugs. However, the actual hit rate of virtual screening is low and most of the predicted drug candidates are false positives.
The research group developed effective filtering algorithms before and after the docking simulations to improve the hit rates. In the pre-docking filtering process, compounds with similar shapes to the known active compounds for each target protein were selected and used for docking simulations. In the post-docking filtering process, the chemicals identified through their docking simulations were evaluated considering the docking energy and the similarity of the protein-ligand interactions with the known active compounds.
The experimental results showed that the virtual screening strategy reached a high hit rate of 18.4%, leading to the identification of seven potential drugs out of the 38 drugs initially selected.
“We plan to conduct further preclinical trials for optimizing drug concentrations as one of the three candidates didn’t resolve the toxicity issues in preclinical trials,” said Woo Dae Jang, one of the researchers from KAIST.
“The most important part of this research is that we developed a platform technology that can rapidly identify novel compounds for COVID-19 treatment. If we use this technology, we will be able to quickly respond to new infectious diseases as well as variants of the coronavirus,” said Distinguished Professor Sang Yup Lee.
This work was supported by the KAIST Mobile Clinic Module Project funded by the Ministry of Science and ICT (MSIT) and the National Research Foundation of Korea (NRF). The National Culture Collection for Pathogens in Korea provided the SARS-CoV-2 (NCCP43326).
-PublicationWoo Dae Jang, Sangeun Jeon, Seungtaek Kim, and Sang Yup Lee. Drugs repurposed for COVID-19 by virtual screening of 6,218 drugs and cell-based assay. Proc. Natl. Acad. Sci. U.S.A. (https://doi/org/10.1073/pnas.2024302118)
-ProfileDistinguished Professor Sang Yup LeeMetabolic &Biomolecular Engineering National Research Laboratoryhttp://mbel.kaist.ac.kr
Department of Chemical and Biomolecular EngineeringKAIST
2021.07.08 View 17292 -
 Natural Rainbow Colorants Microbially Produced
Integrated strategies of systems metabolic engineering and membrane engineering led to the production of natural rainbow colorants comprising seven natural colorants from bacteria for the first time
A research group at KAIST has engineered bacterial strains capable of producing three carotenoids and four violacein derivatives, completing the seven colors in the rainbow spectrum. The research team integrated systems metabolic engineering and membrane engineering strategies for the production of seven natural rainbow colorants in engineered Escherichia coli strains. The strategies will be also useful for the efficient production of other industrially important natural products used in the food, pharmaceutical, and cosmetic industries.
Colorants are widely used in our lives and are directly related to human health when we eat food additives and wear cosmetics. However, most of these colorants are made from petroleum, causing unexpected side effects and health problems. Furthermore, they raise environmental concerns such as water pollution from dyeing fabric in the textiles industry. For these reasons, the demand for the production of natural colorants using microorganisms has increased, but could not be readily realized due to the high cost and low yield of the bioprocesses.
These challenges inspired the metabolic engineers at KAIST including researchers Dr. Dongsoo Yang and Dr. Seon Young Park, and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering. The team reported the study entitled “Production of rainbow colorants by metabolically engineered Escherichia coli” in Advanced Science online on May 5. It was selected as the journal cover of the July 7 issue.
This research reports for the first time the production of rainbow colorants comprising three carotenoids and four violacein derivatives from glucose or glycerol via systems metabolic engineering and membrane engineering. The research group focused on the production of hydrophobic natural colorants useful for lipophilic food and dyeing garments. First, using systems metabolic engineering, which is an integrated technology to engineer the metabolism of a microorganism, three carotenoids comprising astaxanthin (red), -carotene (orange), and zeaxanthin (yellow), and four violacein derivatives comprising proviolacein (green), prodeoxyviolacein (blue), violacein (navy), and deoxyviolacein (purple) could be produced. Thus, the production of natural colorants covering the complete rainbow spectrum was achieved.
When hydrophobic colorants are produced from microorganisms, the colorants are accumulated inside the cell. As the accumulation capacity is limited, the hydrophobic colorants could not be produced with concentrations higher than the limit. In this regard, the researchers engineered the cell morphology and generated inner-membrane vesicles (spherical membranous structures) to increase the intracellular capacity for accumulating the natural colorants. To further promote production, the researchers generated outer-membrane vesicles to secrete the natural colorants, thus succeeding in efficiently producing all of seven rainbow colorants. It was even more impressive that the production of natural green and navy colorants was achieved for the first time.
“The production of the seven natural rainbow colorants that can replace the current petroleum-based synthetic colorants was achieved for the first time,” said Dr. Dongsoo Yang. He explained that another important point of the research is that integrated metabolic engineering strategies developed from this study can be generally applicable for the efficient production of other natural products useful as pharmaceuticals or nutraceuticals. “As maintaining good health in an aging society is becoming increasingly important, we expect that the technology and strategies developed here will play pivotal roles in producing other valuable natural products of medical or nutritional importance,” explained Distinguished Professor Lee.
This work was supported by the "Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01550602)" Rural Development Administration, Republic of Korea.
-Publication:Dongsoo Yang, Seon Young Park, and Sang Yup Lee. Production of rainbow colorants by metabolically engineered Escherichia coli. Advanced Science, 2100743.
-Profile Distinguished Professor Sang Yup LeeMetabolic &Biomolecular Engineering National Research Laboratoryhttp://mbel.kaist.ac.kr
Department of Chemical and Biomolecular EngineeringKAIST
2021.06.09 View 13573
Natural Rainbow Colorants Microbially Produced
Integrated strategies of systems metabolic engineering and membrane engineering led to the production of natural rainbow colorants comprising seven natural colorants from bacteria for the first time
A research group at KAIST has engineered bacterial strains capable of producing three carotenoids and four violacein derivatives, completing the seven colors in the rainbow spectrum. The research team integrated systems metabolic engineering and membrane engineering strategies for the production of seven natural rainbow colorants in engineered Escherichia coli strains. The strategies will be also useful for the efficient production of other industrially important natural products used in the food, pharmaceutical, and cosmetic industries.
Colorants are widely used in our lives and are directly related to human health when we eat food additives and wear cosmetics. However, most of these colorants are made from petroleum, causing unexpected side effects and health problems. Furthermore, they raise environmental concerns such as water pollution from dyeing fabric in the textiles industry. For these reasons, the demand for the production of natural colorants using microorganisms has increased, but could not be readily realized due to the high cost and low yield of the bioprocesses.
These challenges inspired the metabolic engineers at KAIST including researchers Dr. Dongsoo Yang and Dr. Seon Young Park, and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering. The team reported the study entitled “Production of rainbow colorants by metabolically engineered Escherichia coli” in Advanced Science online on May 5. It was selected as the journal cover of the July 7 issue.
This research reports for the first time the production of rainbow colorants comprising three carotenoids and four violacein derivatives from glucose or glycerol via systems metabolic engineering and membrane engineering. The research group focused on the production of hydrophobic natural colorants useful for lipophilic food and dyeing garments. First, using systems metabolic engineering, which is an integrated technology to engineer the metabolism of a microorganism, three carotenoids comprising astaxanthin (red), -carotene (orange), and zeaxanthin (yellow), and four violacein derivatives comprising proviolacein (green), prodeoxyviolacein (blue), violacein (navy), and deoxyviolacein (purple) could be produced. Thus, the production of natural colorants covering the complete rainbow spectrum was achieved.
When hydrophobic colorants are produced from microorganisms, the colorants are accumulated inside the cell. As the accumulation capacity is limited, the hydrophobic colorants could not be produced with concentrations higher than the limit. In this regard, the researchers engineered the cell morphology and generated inner-membrane vesicles (spherical membranous structures) to increase the intracellular capacity for accumulating the natural colorants. To further promote production, the researchers generated outer-membrane vesicles to secrete the natural colorants, thus succeeding in efficiently producing all of seven rainbow colorants. It was even more impressive that the production of natural green and navy colorants was achieved for the first time.
“The production of the seven natural rainbow colorants that can replace the current petroleum-based synthetic colorants was achieved for the first time,” said Dr. Dongsoo Yang. He explained that another important point of the research is that integrated metabolic engineering strategies developed from this study can be generally applicable for the efficient production of other natural products useful as pharmaceuticals or nutraceuticals. “As maintaining good health in an aging society is becoming increasingly important, we expect that the technology and strategies developed here will play pivotal roles in producing other valuable natural products of medical or nutritional importance,” explained Distinguished Professor Lee.
This work was supported by the "Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01550602)" Rural Development Administration, Republic of Korea.
-Publication:Dongsoo Yang, Seon Young Park, and Sang Yup Lee. Production of rainbow colorants by metabolically engineered Escherichia coli. Advanced Science, 2100743.
-Profile Distinguished Professor Sang Yup LeeMetabolic &Biomolecular Engineering National Research Laboratoryhttp://mbel.kaist.ac.kr
Department of Chemical and Biomolecular EngineeringKAIST
2021.06.09 View 13573 -
 Prof. Sang Yup Lee Elected as a Foreign Member of the Royal Society
Vice President for Research Distinguished Professor Sang Yup Lee was elected as a foreign member of the Royal Society in the UK. On May 6, the Society announced the list of distinguished new 52 fellows and 10 foreign members who achieved exceptional contributions to science. Professor Lee and Professor V. Narry Kim from Seoul National University are the first foreign members ever elected from Korea.
The Royal Society, established in 1660, is one of the most prestigious national science academies and a fellowship of 1,600 of the world’s most eminent scientists. From Newton to Darwin, Einstein, Hawking, and beyond, pioneers and paragons in their fields are elected by their peers. To date, there are 280 Nobel prize winners among the fellows.
Distinguished Professor Lee from the Department of Chemical and Biomolecular Engineering at KAIST is one of the Highly Cited Researchers (HCRs) who pioneered systems metabolic engineering and developed various micro-organisms for producing a wide range of fuels, chemicals, materials, and natural compounds.
His seminal scholarship and research career have already been recognized worldwide. He is the first Korean ever elected into the National Academy of Inventors (NAI) in the US and one of 13 scholars elected as an International Member of both the National Academy of Sciences (NAS) and the National Academy of Engineering (NAE) in the US. With this fellowship, he added one more accolade of being the first non-US and British Commonwealth scientist elected into the three most prestigious science academies: the NAS, the NAE, and the Royal Society.
2021.05.07 View 14042
Prof. Sang Yup Lee Elected as a Foreign Member of the Royal Society
Vice President for Research Distinguished Professor Sang Yup Lee was elected as a foreign member of the Royal Society in the UK. On May 6, the Society announced the list of distinguished new 52 fellows and 10 foreign members who achieved exceptional contributions to science. Professor Lee and Professor V. Narry Kim from Seoul National University are the first foreign members ever elected from Korea.
The Royal Society, established in 1660, is one of the most prestigious national science academies and a fellowship of 1,600 of the world’s most eminent scientists. From Newton to Darwin, Einstein, Hawking, and beyond, pioneers and paragons in their fields are elected by their peers. To date, there are 280 Nobel prize winners among the fellows.
Distinguished Professor Lee from the Department of Chemical and Biomolecular Engineering at KAIST is one of the Highly Cited Researchers (HCRs) who pioneered systems metabolic engineering and developed various micro-organisms for producing a wide range of fuels, chemicals, materials, and natural compounds.
His seminal scholarship and research career have already been recognized worldwide. He is the first Korean ever elected into the National Academy of Inventors (NAI) in the US and one of 13 scholars elected as an International Member of both the National Academy of Sciences (NAS) and the National Academy of Engineering (NAE) in the US. With this fellowship, he added one more accolade of being the first non-US and British Commonwealth scientist elected into the three most prestigious science academies: the NAS, the NAE, and the Royal Society.
2021.05.07 View 14042 -
 Distinguished Professor Sang Yup Lee Honored with Charles D. Scott Award
Vice President for Research Sang Yup Lee received the 2021 Charles D. Scott Award from the Society for Industrial Microbiology and Biotechnology. Distinguished Professor Lee from the Department of Chemical and Biomolecular Engineering at KAIST is the first Asian awardee.
The Charles D. Scott Award, initiated in 1995, recognizes individuals who have made significant contributions to enable and further the use of biotechnology to produce fuels and chemicals. The award is named in honor of Dr. Charles D. Scott, who founded the Symposium on Biomaterials, Fuels, and Chemicals and chaired the conference for its first ten years.
Professor Lee has pioneered systems metabolic engineering and developed various micro-organisms capable of producing a wide range of fuels, chemicals, materials, and natural compounds, many of them for the first time. Some of the breakthroughs include the microbial production of gasoline, diacids, diamines, PLA and PLGA polymers, and several natural products.
More recently, his team has developed a microbial strain capable of the mass production of succinic acid, a monomer for manufacturing polyester, with the highest production efficiency to date, as well as a Corynebacterium glutamicum strain capable of producing high-level glutaric acid. They also engineered for the first time a bacterium capable of producing carminic acid, a natural red colorant that is widely used for food and cosmetics.
Professor Lee is one of the Highly Cited Researchers (HCR), ranked in the top 1% by citations in their field by Clarivate Analytics for four consecutive years from 2017. He is the first Korean fellow ever elected into the National Academy of Inventors in the US and one of 13 scholars elected as an International Member of both the National Academy of Sciences and the National Academy of Engineering in the USA.
The awards ceremony will take place during the Symposium on Biomaterials, Fuels, and Chemicals held online from April 26.
2021.04.27 View 11829
Distinguished Professor Sang Yup Lee Honored with Charles D. Scott Award
Vice President for Research Sang Yup Lee received the 2021 Charles D. Scott Award from the Society for Industrial Microbiology and Biotechnology. Distinguished Professor Lee from the Department of Chemical and Biomolecular Engineering at KAIST is the first Asian awardee.
The Charles D. Scott Award, initiated in 1995, recognizes individuals who have made significant contributions to enable and further the use of biotechnology to produce fuels and chemicals. The award is named in honor of Dr. Charles D. Scott, who founded the Symposium on Biomaterials, Fuels, and Chemicals and chaired the conference for its first ten years.
Professor Lee has pioneered systems metabolic engineering and developed various micro-organisms capable of producing a wide range of fuels, chemicals, materials, and natural compounds, many of them for the first time. Some of the breakthroughs include the microbial production of gasoline, diacids, diamines, PLA and PLGA polymers, and several natural products.
More recently, his team has developed a microbial strain capable of the mass production of succinic acid, a monomer for manufacturing polyester, with the highest production efficiency to date, as well as a Corynebacterium glutamicum strain capable of producing high-level glutaric acid. They also engineered for the first time a bacterium capable of producing carminic acid, a natural red colorant that is widely used for food and cosmetics.
Professor Lee is one of the Highly Cited Researchers (HCR), ranked in the top 1% by citations in their field by Clarivate Analytics for four consecutive years from 2017. He is the first Korean fellow ever elected into the National Academy of Inventors in the US and one of 13 scholars elected as an International Member of both the National Academy of Sciences and the National Academy of Engineering in the USA.
The awards ceremony will take place during the Symposium on Biomaterials, Fuels, and Chemicals held online from April 26.
2021.04.27 View 11829 -
 Microbial Production of a Natural Red Colorant Carminic Acid
Metabolic engineering and computer-simulated enzyme engineering led to the production of carminic acid, a natural red colorant, from bacteria for the first time
A research group at KAIST has engineered a bacterium capable of producing a natural red colorant, carminic acid, which is widely used for food and cosmetics. The research team reported the complete biosynthesis of carminic acid from glucose in engineered Escherichia coli. The strategies will be useful for the design and construction of biosynthetic pathways involving unknown enzymes and consequently the production of diverse industrially important natural products for the food, pharmaceutical, and cosmetic industries.
Carminic acid is a natural red colorant widely being used for products such as strawberry milk and lipstick. However, carminic acid has been produced by farming cochineals, a scale insect which only grows in the region around Peru and Canary Islands, followed by complicated multi-step purification processes. Moreover, carminic acid often contains protein contaminants that cause allergies so many people are unwilling to consume products made of insect-driven colorants. On that account, manufacturers around the world are using alternative red colorants despite the fact that carminic acid is one of the most stable natural red colorants.
These challenges inspired the metabolic engineering research group at KAIST to address this issue. Its members include postdoctoral researchers Dongsoo Yang and Woo Dae Jang, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering. This study entitled “Production of carminic acid by metabolically engineered Escherichia coli” was published online in the Journal of the American Chemical Society (JACS) on April 2.
This research reports for the first time the development of a bacterial strain capable of producing carminic acid from glucose via metabolic engineering and computer simulation-assisted enzyme engineering. The research group optimized the type II polyketide synthase machinery to efficiently produce the precursor of carminic acid, flavokermesic acid.
Since the enzymes responsible for the remaining two reactions were neither discovered nor functional, biochemical reaction analysis was performed to identify enzymes that can convert flavokermesic acid into carminic acid. Then, homology modeling and docking simulations were performed to enhance the activities of the two identified enzymes. The team could confirm that the final engineered strain could produce carminic acid directly from glucose. The C-glucosyltransferase developed in this study was found to be generally applicable for other natural products as showcased by the successful production of an additional product, aloesin, which is found in aloe leaves.
“The most important part of this research is that unknown enzymes for the production of target natural products were identified and improved by biochemical reaction analyses and computer simulation-assisted enzyme engineering,” says Dr. Dongsoo Yang. He explained the development of a generally applicable C-glucosyltransferase is also useful since C-glucosylation is a relatively unexplored reaction in bacteria including Escherichia coli. Using the C-glucosyltransferase developed in this study, both carminic acid and aloesin were successfully produced from glucose.
“A sustainable and insect-free method of producing carminic acid was achieved for the first time in this study. Unknown or inefficient enzymes have always been a major problem in natural product biosynthesis, and here we suggest one effective solution for solving this problem. As maintaining good health in the aging society is becoming increasingly important, we expect that the technology and strategies developed here will play pivotal roles in producing other valuable natural products of medical or nutritional importance,” said Distinguished Professor Sang Yup Lee.
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries of the Ministry of Science and ICT (MSIT) through the National Research Foundation (NRF) of Korea and the KAIST Cross-Generation Collaborative Lab project; Sang Yup Lee and Dongsoo Yang were also supported by Novo Nordisk Foundation in Denmark.
Publication:
Dongsoo Yang, Woo Dae Jang, and Sang Yup Lee. Production of carminic acid by metabolically engineered Escherichia coli. at the Journal of the American Chemical Society. https://doi.org.10.1021/jacs.0c12406
Profile:
Sang Yup Lee, PhD
Distinguished Professor
leesy@kaist.ac.kr
http://mbel.kaist.ac.kr
Metabolic &Biomolecular Engineering National Research Laboratory
Department of Chemical and Biomolecular Engineering
KAIST
2021.04.06 View 14743
Microbial Production of a Natural Red Colorant Carminic Acid
Metabolic engineering and computer-simulated enzyme engineering led to the production of carminic acid, a natural red colorant, from bacteria for the first time
A research group at KAIST has engineered a bacterium capable of producing a natural red colorant, carminic acid, which is widely used for food and cosmetics. The research team reported the complete biosynthesis of carminic acid from glucose in engineered Escherichia coli. The strategies will be useful for the design and construction of biosynthetic pathways involving unknown enzymes and consequently the production of diverse industrially important natural products for the food, pharmaceutical, and cosmetic industries.
Carminic acid is a natural red colorant widely being used for products such as strawberry milk and lipstick. However, carminic acid has been produced by farming cochineals, a scale insect which only grows in the region around Peru and Canary Islands, followed by complicated multi-step purification processes. Moreover, carminic acid often contains protein contaminants that cause allergies so many people are unwilling to consume products made of insect-driven colorants. On that account, manufacturers around the world are using alternative red colorants despite the fact that carminic acid is one of the most stable natural red colorants.
These challenges inspired the metabolic engineering research group at KAIST to address this issue. Its members include postdoctoral researchers Dongsoo Yang and Woo Dae Jang, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering. This study entitled “Production of carminic acid by metabolically engineered Escherichia coli” was published online in the Journal of the American Chemical Society (JACS) on April 2.
This research reports for the first time the development of a bacterial strain capable of producing carminic acid from glucose via metabolic engineering and computer simulation-assisted enzyme engineering. The research group optimized the type II polyketide synthase machinery to efficiently produce the precursor of carminic acid, flavokermesic acid.
Since the enzymes responsible for the remaining two reactions were neither discovered nor functional, biochemical reaction analysis was performed to identify enzymes that can convert flavokermesic acid into carminic acid. Then, homology modeling and docking simulations were performed to enhance the activities of the two identified enzymes. The team could confirm that the final engineered strain could produce carminic acid directly from glucose. The C-glucosyltransferase developed in this study was found to be generally applicable for other natural products as showcased by the successful production of an additional product, aloesin, which is found in aloe leaves.
“The most important part of this research is that unknown enzymes for the production of target natural products were identified and improved by biochemical reaction analyses and computer simulation-assisted enzyme engineering,” says Dr. Dongsoo Yang. He explained the development of a generally applicable C-glucosyltransferase is also useful since C-glucosylation is a relatively unexplored reaction in bacteria including Escherichia coli. Using the C-glucosyltransferase developed in this study, both carminic acid and aloesin were successfully produced from glucose.
“A sustainable and insect-free method of producing carminic acid was achieved for the first time in this study. Unknown or inefficient enzymes have always been a major problem in natural product biosynthesis, and here we suggest one effective solution for solving this problem. As maintaining good health in the aging society is becoming increasingly important, we expect that the technology and strategies developed here will play pivotal roles in producing other valuable natural products of medical or nutritional importance,” said Distinguished Professor Sang Yup Lee.
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries of the Ministry of Science and ICT (MSIT) through the National Research Foundation (NRF) of Korea and the KAIST Cross-Generation Collaborative Lab project; Sang Yup Lee and Dongsoo Yang were also supported by Novo Nordisk Foundation in Denmark.
Publication:
Dongsoo Yang, Woo Dae Jang, and Sang Yup Lee. Production of carminic acid by metabolically engineered Escherichia coli. at the Journal of the American Chemical Society. https://doi.org.10.1021/jacs.0c12406
Profile:
Sang Yup Lee, PhD
Distinguished Professor
leesy@kaist.ac.kr
http://mbel.kaist.ac.kr
Metabolic &Biomolecular Engineering National Research Laboratory
Department of Chemical and Biomolecular Engineering
KAIST
2021.04.06 View 14743 -
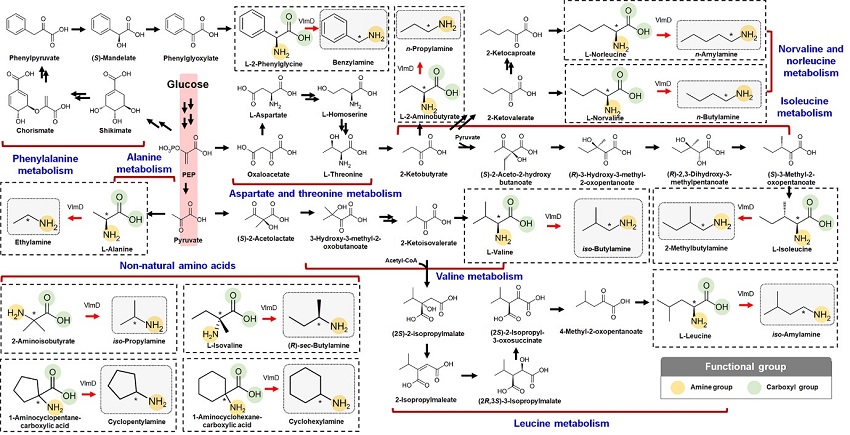 Expanding the Biosynthetic Pathway via Retrobiosynthesis
- Researchers reports a new strategy for the microbial production of multiple short-chain primary amines via retrobiosynthesis. -
KAIST metabolic engineers presented the bio-based production of multiple short-chain primary amines that have a wide range of applications in chemical industries for the first time. The research team led by Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering designed the novel biosynthetic pathways for short-chain primary amines by combining retrobiosynthesis and a precursor selection step.
The research team verified the newly designed pathways by confirming the in vivo production of 10 short-chain primary amines by supplying the precursors. Furthermore, the platform Escherichia coli strains were metabolically engineered to produce three proof-of-concept short-chain primary amines from glucose, demonstrating the possibility of the bio-based production of diverse short-chain primary amines from renewable resources. The research team said this study expands the strategy of systematically designing biosynthetic pathways for the production of a group of related chemicals as demonstrated by multiple short-chain primary amines as examples.
Currently, most of the industrial chemicals used in our daily lives are produced with petroleum-based products. However, there are several serious issues with the petroleum industry such as the depletion of fossil fuel reserves and environmental problems including global warming. To solve these problems, the sustainable production of industrial chemicals and materials is being explored with microorganisms as cell factories and renewable non-food biomass as raw materials for alternative to petroleum-based products. The engineering of these microorganisms has increasingly become more efficient and effective with the help of systems metabolic engineering – a practice of engineering the metabolism of a living organism toward the production of a desired metabolite. In this regard, the number of chemicals produced using biomass as a raw material has substantially increased.
Although the scope of chemicals that are producible using microorganisms continues to expand through advances in systems metabolic engineering, the biological production of short-chain primary amines has not yet been reported despite their industrial importance. Short-chain primary amines are the chemicals that have an alkyl or aryl group in the place of a hydrogen atom in ammonia with carbon chain lengths ranging from C1 to C7. Short-chain primary amines have a wide range of applications in chemical industries, for example, as a precursor for pharmaceuticals (e.g., antidiabetic and antihypertensive drugs), agrochemicals (e.g., herbicides, fungicides and insecticides), solvents, and vulcanization accelerators for rubber and plasticizers. The market size of short-chain primary amines was estimated to be more than 4 billion US dollars in 2014.
The main reason why the bio-based production of short-chain primary amines was not yet possible was due to their unknown biosynthetic pathways. Therefore, the team designed synthetic biosynthetic pathways for short-chain primary amines by combining retrobiosynthesis and a precursor selection step. The retrobiosynthesis allowed the systematic design of a biosynthetic pathway for short-chain primary amines by using a set of biochemical reaction rules that describe chemical transformation patterns between a substrate and product molecules at an atomic level.
These multiple precursors predicted for the possible biosynthesis of each short-chain primary amine were sequentially narrowed down by using the precursor selection step for efficient metabolic engineering experiments.
“Our research demonstrates the possibility of the renewable production of short-chain primary amines for the first time. We are planning to increase production efficiencies of short-chain primary amines. We believe that our study will play an important role in the development of sustainable and eco-friendly bio-based industries and the reorganization of the chemical industry, which is mandatory for solving the environmental problems threating the survival of mankind,” said Professor Lee.
This paper titled “Microbial production of multiple short-chain primary amines via retrobiosynthesis” was published in Nature Communications. This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science and ICT through the National Research Foundation (NRF) of Korea.
-Publication
Dong In Kim, Tong Un Chae, Hyun Uk Kim, Woo Dae Jang, and Sang Yup Lee. Microbial production of multiple short-chain primary amines via retrobiosynthesis. Nature Communications ( https://www.nature.com/articles/s41467-020-20423-6)
-Profile
Distinguished Professor Sang Yup Lee
leesy@kaist.ac.kr
Metabolic &Biomolecular Engineering National Research Laboratory
http://mbel.kaist.ac.kr
Department of Chemical and Biomolecular Engineering
KAIST
2021.01.14 View 14568
Expanding the Biosynthetic Pathway via Retrobiosynthesis
- Researchers reports a new strategy for the microbial production of multiple short-chain primary amines via retrobiosynthesis. -
KAIST metabolic engineers presented the bio-based production of multiple short-chain primary amines that have a wide range of applications in chemical industries for the first time. The research team led by Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering designed the novel biosynthetic pathways for short-chain primary amines by combining retrobiosynthesis and a precursor selection step.
The research team verified the newly designed pathways by confirming the in vivo production of 10 short-chain primary amines by supplying the precursors. Furthermore, the platform Escherichia coli strains were metabolically engineered to produce three proof-of-concept short-chain primary amines from glucose, demonstrating the possibility of the bio-based production of diverse short-chain primary amines from renewable resources. The research team said this study expands the strategy of systematically designing biosynthetic pathways for the production of a group of related chemicals as demonstrated by multiple short-chain primary amines as examples.
Currently, most of the industrial chemicals used in our daily lives are produced with petroleum-based products. However, there are several serious issues with the petroleum industry such as the depletion of fossil fuel reserves and environmental problems including global warming. To solve these problems, the sustainable production of industrial chemicals and materials is being explored with microorganisms as cell factories and renewable non-food biomass as raw materials for alternative to petroleum-based products. The engineering of these microorganisms has increasingly become more efficient and effective with the help of systems metabolic engineering – a practice of engineering the metabolism of a living organism toward the production of a desired metabolite. In this regard, the number of chemicals produced using biomass as a raw material has substantially increased.
Although the scope of chemicals that are producible using microorganisms continues to expand through advances in systems metabolic engineering, the biological production of short-chain primary amines has not yet been reported despite their industrial importance. Short-chain primary amines are the chemicals that have an alkyl or aryl group in the place of a hydrogen atom in ammonia with carbon chain lengths ranging from C1 to C7. Short-chain primary amines have a wide range of applications in chemical industries, for example, as a precursor for pharmaceuticals (e.g., antidiabetic and antihypertensive drugs), agrochemicals (e.g., herbicides, fungicides and insecticides), solvents, and vulcanization accelerators for rubber and plasticizers. The market size of short-chain primary amines was estimated to be more than 4 billion US dollars in 2014.
The main reason why the bio-based production of short-chain primary amines was not yet possible was due to their unknown biosynthetic pathways. Therefore, the team designed synthetic biosynthetic pathways for short-chain primary amines by combining retrobiosynthesis and a precursor selection step. The retrobiosynthesis allowed the systematic design of a biosynthetic pathway for short-chain primary amines by using a set of biochemical reaction rules that describe chemical transformation patterns between a substrate and product molecules at an atomic level.
These multiple precursors predicted for the possible biosynthesis of each short-chain primary amine were sequentially narrowed down by using the precursor selection step for efficient metabolic engineering experiments.
“Our research demonstrates the possibility of the renewable production of short-chain primary amines for the first time. We are planning to increase production efficiencies of short-chain primary amines. We believe that our study will play an important role in the development of sustainable and eco-friendly bio-based industries and the reorganization of the chemical industry, which is mandatory for solving the environmental problems threating the survival of mankind,” said Professor Lee.
This paper titled “Microbial production of multiple short-chain primary amines via retrobiosynthesis” was published in Nature Communications. This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science and ICT through the National Research Foundation (NRF) of Korea.
-Publication
Dong In Kim, Tong Un Chae, Hyun Uk Kim, Woo Dae Jang, and Sang Yup Lee. Microbial production of multiple short-chain primary amines via retrobiosynthesis. Nature Communications ( https://www.nature.com/articles/s41467-020-20423-6)
-Profile
Distinguished Professor Sang Yup Lee
leesy@kaist.ac.kr
Metabolic &Biomolecular Engineering National Research Laboratory
http://mbel.kaist.ac.kr
Department of Chemical and Biomolecular Engineering
KAIST
2021.01.14 View 14568 -
 DeepTFactor Predicts Transcription Factors
A deep learning-based tool predicts transcription factors using protein sequences as inputs
A joint research team from KAIST and UCSD has developed a deep neural network named DeepTFactor that predicts transcription factors from protein sequences. DeepTFactor will serve as a useful tool for understanding the regulatory systems of organisms, accelerating the use of deep learning for solving biological problems.
A transcription factor is a protein that specifically binds to DNA sequences to control the transcription initiation. Analyzing transcriptional regulation enables the understanding of how organisms control gene expression in response to genetic or environmental changes. In this regard, finding the transcription factor of an organism is the first step in the analysis of the transcriptional regulatory system of an organism.
Previously, transcription factors have been predicted by analyzing sequence homology with already characterized transcription factors or by data-driven approaches such as machine learning. Conventional machine learning models require a rigorous feature selection process that relies on domain expertise such as calculating the physicochemical properties of molecules or analyzing the homology of biological sequences. Meanwhile, deep learning can inherently learn latent features for the specific task.
A joint research team comprised of Ph.D. candidate Gi Bae Kim and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST, and Ye Gao and Professor Bernhard O. Palsson of the Department of Biochemical Engineering at UCSD reported a deep learning-based tool for the prediction of transcription factors. Their research paper “DeepTFactor: A deep learning-based tool for the prediction of transcription factors” was published online in PNAS.
Their article reports the development of DeepTFactor, a deep learning-based tool that predicts whether a given protein sequence is a transcription factor using three parallel convolutional neural networks. The joint research team predicted 332 transcription factors of Escherichia coli K-12 MG1655 using DeepTFactor and the performance of DeepTFactor by experimentally confirming the genome-wide binding sites of three predicted transcription factors (YqhC, YiaU, and YahB).
The joint research team further used a saliency method to understand the reasoning process of DeepTFactor. The researchers confirmed that even though information on the DNA binding domains of the transcription factor was not explicitly given the training process, DeepTFactor implicitly learned and used them for prediction. Unlike previous transcription factor prediction tools that were developed only for protein sequences of specific organisms, DeepTFactor is expected to be used in the analysis of the transcription systems of all organisms at a high level of performance.
Distinguished Professor Sang Yup Lee said, “DeepTFactor can be used to discover unknown transcription factors from numerous protein sequences that have not yet been characterized. It is expected that DeepTFactor will serve as an important tool for analyzing the regulatory systems of organisms of interest.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science and ICT through the National Research Foundation of Korea.
-Publication
Gi Bae Kim, Ye Gao, Bernhard O. Palsson, and Sang Yup Lee. DeepTFactor: A deep learning-based tool for the prediction of transcription factors. (https://doi.org/10.1073/pnas202117118)
-Profile
Distinguished Professor Sang Yup Lee
leesy@kaist.ac.kr
Metabolic &Biomolecular Engineering National Research Laboratory
http://mbel.kaist.ac.kr
Department of Chemical and Biomolecular Engineering
KAIST
2021.01.05 View 11522
DeepTFactor Predicts Transcription Factors
A deep learning-based tool predicts transcription factors using protein sequences as inputs
A joint research team from KAIST and UCSD has developed a deep neural network named DeepTFactor that predicts transcription factors from protein sequences. DeepTFactor will serve as a useful tool for understanding the regulatory systems of organisms, accelerating the use of deep learning for solving biological problems.
A transcription factor is a protein that specifically binds to DNA sequences to control the transcription initiation. Analyzing transcriptional regulation enables the understanding of how organisms control gene expression in response to genetic or environmental changes. In this regard, finding the transcription factor of an organism is the first step in the analysis of the transcriptional regulatory system of an organism.
Previously, transcription factors have been predicted by analyzing sequence homology with already characterized transcription factors or by data-driven approaches such as machine learning. Conventional machine learning models require a rigorous feature selection process that relies on domain expertise such as calculating the physicochemical properties of molecules or analyzing the homology of biological sequences. Meanwhile, deep learning can inherently learn latent features for the specific task.
A joint research team comprised of Ph.D. candidate Gi Bae Kim and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST, and Ye Gao and Professor Bernhard O. Palsson of the Department of Biochemical Engineering at UCSD reported a deep learning-based tool for the prediction of transcription factors. Their research paper “DeepTFactor: A deep learning-based tool for the prediction of transcription factors” was published online in PNAS.
Their article reports the development of DeepTFactor, a deep learning-based tool that predicts whether a given protein sequence is a transcription factor using three parallel convolutional neural networks. The joint research team predicted 332 transcription factors of Escherichia coli K-12 MG1655 using DeepTFactor and the performance of DeepTFactor by experimentally confirming the genome-wide binding sites of three predicted transcription factors (YqhC, YiaU, and YahB).
The joint research team further used a saliency method to understand the reasoning process of DeepTFactor. The researchers confirmed that even though information on the DNA binding domains of the transcription factor was not explicitly given the training process, DeepTFactor implicitly learned and used them for prediction. Unlike previous transcription factor prediction tools that were developed only for protein sequences of specific organisms, DeepTFactor is expected to be used in the analysis of the transcription systems of all organisms at a high level of performance.
Distinguished Professor Sang Yup Lee said, “DeepTFactor can be used to discover unknown transcription factors from numerous protein sequences that have not yet been characterized. It is expected that DeepTFactor will serve as an important tool for analyzing the regulatory systems of organisms of interest.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science and ICT through the National Research Foundation of Korea.
-Publication
Gi Bae Kim, Ye Gao, Bernhard O. Palsson, and Sang Yup Lee. DeepTFactor: A deep learning-based tool for the prediction of transcription factors. (https://doi.org/10.1073/pnas202117118)
-Profile
Distinguished Professor Sang Yup Lee
leesy@kaist.ac.kr
Metabolic &Biomolecular Engineering National Research Laboratory
http://mbel.kaist.ac.kr
Department of Chemical and Biomolecular Engineering
KAIST
2021.01.05 View 11522 -
 A Comprehensive Review of Biosynthesis of Inorganic Nanomaterials Using Microorganisms and Bacteriophages
There are diverse methods for producing numerous inorganic nanomaterials involving many experimental variables. Among the numerous possible matches, finding the best pair for synthesizing in an environmentally friendly way has been a longstanding challenge for researchers and industries.
A KAIST bioprocess engineering research team led by Distinguished Professor Sang Yup Lee conducted a summary of 146 biosynthesized single and multi-element inorganic nanomaterials covering 55 elements in the periodic table synthesized using wild-type and genetically engineered microorganisms. Their research highlights the diverse applications of biogenic nanomaterials and gives strategies for improving the biosynthesis of nanomaterials in terms of their producibility, crystallinity, size, and shape.
The research team described a 10-step flow chart for developing the biosynthesis of inorganic nanomaterials using microorganisms and bacteriophages. The research was published at Nature Review Chemistry as a cover and hero paper on December 3.
“We suggest general strategies for microbial nanomaterial biosynthesis via a step-by-step flow chart and give our perspectives on the future of nanomaterial biosynthesis and applications. This flow chart will serve as a general guide for those wishing to prepare biosynthetic inorganic nanomaterials using microbial cells,” explained Dr.Yoojin Choi, a co-author of this research.
Most inorganic nanomaterials are produced using physical and chemical methods and biological synthesis has been gaining more and more attention. However, conventional synthesis processes have drawbacks in terms of high energy consumption and non-environmentally friendly processes. Meanwhile, microorganisms such as microalgae, yeasts, fungi, bacteria, and even viruses can be utilized as biofactories to produce single and multi-element inorganic nanomaterials under mild conditions.
After conducting a massive survey, the research team summed up that the development of genetically engineered microorganisms with increased inorganic-ion-binding affinity, inorganic-ion-reduction ability, and nanomaterial biosynthetic efficiency has enabled the synthesis of many inorganic nanomaterials.
Among the strategies, the team introduced their analysis of a Pourbaix diagram for controlling the size and morphology of a product. The research team said this Pourbaix diagram analysis can be widely employed for biosynthesizing new nanomaterials with industrial applications.Professor Sang Yup Lee added, “This research provides extensive information and perspectives on the biosynthesis of diverse inorganic nanomaterials using microorganisms and bacteriophages and their applications. We expect that biosynthetic inorganic nanomaterials will find more diverse and innovative applications across diverse fields of science and technology.”
Dr. Choi started this research in 2018 and her interview about completing this extensive research was featured in an article at Nature Career article on December 4.
-ProfileDistinguished Professor Sang Yup Lee leesy@kaist.ac.krMetabolic &Biomolecular Engineering National Research Laboratoryhttp://mbel.kaist.ac.krDepartment of Chemical and Biomolecular EngineeringKAIST
2020.12.07 View 13473
A Comprehensive Review of Biosynthesis of Inorganic Nanomaterials Using Microorganisms and Bacteriophages
There are diverse methods for producing numerous inorganic nanomaterials involving many experimental variables. Among the numerous possible matches, finding the best pair for synthesizing in an environmentally friendly way has been a longstanding challenge for researchers and industries.
A KAIST bioprocess engineering research team led by Distinguished Professor Sang Yup Lee conducted a summary of 146 biosynthesized single and multi-element inorganic nanomaterials covering 55 elements in the periodic table synthesized using wild-type and genetically engineered microorganisms. Their research highlights the diverse applications of biogenic nanomaterials and gives strategies for improving the biosynthesis of nanomaterials in terms of their producibility, crystallinity, size, and shape.
The research team described a 10-step flow chart for developing the biosynthesis of inorganic nanomaterials using microorganisms and bacteriophages. The research was published at Nature Review Chemistry as a cover and hero paper on December 3.
“We suggest general strategies for microbial nanomaterial biosynthesis via a step-by-step flow chart and give our perspectives on the future of nanomaterial biosynthesis and applications. This flow chart will serve as a general guide for those wishing to prepare biosynthetic inorganic nanomaterials using microbial cells,” explained Dr.Yoojin Choi, a co-author of this research.
Most inorganic nanomaterials are produced using physical and chemical methods and biological synthesis has been gaining more and more attention. However, conventional synthesis processes have drawbacks in terms of high energy consumption and non-environmentally friendly processes. Meanwhile, microorganisms such as microalgae, yeasts, fungi, bacteria, and even viruses can be utilized as biofactories to produce single and multi-element inorganic nanomaterials under mild conditions.
After conducting a massive survey, the research team summed up that the development of genetically engineered microorganisms with increased inorganic-ion-binding affinity, inorganic-ion-reduction ability, and nanomaterial biosynthetic efficiency has enabled the synthesis of many inorganic nanomaterials.
Among the strategies, the team introduced their analysis of a Pourbaix diagram for controlling the size and morphology of a product. The research team said this Pourbaix diagram analysis can be widely employed for biosynthesizing new nanomaterials with industrial applications.Professor Sang Yup Lee added, “This research provides extensive information and perspectives on the biosynthesis of diverse inorganic nanomaterials using microorganisms and bacteriophages and their applications. We expect that biosynthetic inorganic nanomaterials will find more diverse and innovative applications across diverse fields of science and technology.”
Dr. Choi started this research in 2018 and her interview about completing this extensive research was featured in an article at Nature Career article on December 4.
-ProfileDistinguished Professor Sang Yup Lee leesy@kaist.ac.krMetabolic &Biomolecular Engineering National Research Laboratoryhttp://mbel.kaist.ac.krDepartment of Chemical and Biomolecular EngineeringKAIST
2020.12.07 View 13473