research
-
 KAIST Discovers Protein Switch that Turns Anti-Viral Immune Response On and Off
Even after the COVID-19 pandemic, various new infectious diseases continue to emerge, posing ongoing viral threats that demand robust and sustained immune defenses. However, excessive immune reactions can also harm body tissues, causing significant health issues. KAIST and an international research team have discovered a critical protein that acts as a 'switch' regulating immune responses to viruses. This breakthrough is expected to lay the groundwork for future infectious disease responses and autoimmune disease treatment strategies.
KAIST (President Kwang-Hyung Lee) announced on May 14 that a joint research team led by Professor Yoosik Kim from the Department of Chemical and Biomolecular Engineering at KAIST and Professor Seunghee Cha from University of Florida has discovered the mechanism by which double-stranded RNA derived from mitochondria amplifies immune responses. They identified the protein SLIRP as an 'immune switch' that regulates this process, playing a crucial role in both viral infections and autoimmune diseases.
< (From left) Master's candidate Yewon Yang, Professor Yoosik Kim and Ph.D. candidate Doyeong Ku of the Department of Chemical and Biomolecular Engineering >
Autoimmune diseases arise when the immune system fails to differentiate between external pathogens and the body's own molecules, leading to self-directed attacks. Despite extensive research, the precise causes of excessive inflammatory conditions like Sjögren’s syndrome and systemic lupus erythematosus remain unclear, and effective treatments are still limited.
To uncover the molecular mechanisms driving immune hyperactivation and to identify potential regulatory factors, the research team led by Professor Yoosik Kim focused on mitochondrial double-stranded RNA (mt-dsRNA), a genetic immunogenic material produced within cellular organelles. Since mt-dsRNA structurally resembles viral RNA, it can mistakenly trigger immune responses even in the absence of an actual viral infection.
The team discovered that SLIRP, a key regulator of mt-dsRNA, amplifies immune responses by stabilizing the RNA. They confirmed that SLIRP expression increases in experimental models simulating the tissues of autoimmune disease patients and viral infections. Conversely, suppressing SLIRP significantly reduced the immune response, underscoring its role as a critical factor in immune amplification.
This study also demonstrated the dual function of SLIRP in different contexts. In cells infected with human beta coronavirus OC43 and encephalomyocarditis virus (EMCV), SLIRP suppression led to reduced antiviral responses and increased viral replication. Meanwhile, in the blood and salivary gland cells of Sjögren’s syndrome patients, where both SLIRP and mt-dsRNA levels were elevated, suppressing SLIRP alleviated the abnormal immune response.
These findings highlight SLIRP as a key molecular switch that regulates immune responses in both infections and autoimmune diseases.
< Figure 1. Schematic diagram of antiviral signal amplification by SLIRP: SLIRP-based mt-dsRNA induction, cytoplasmic accumulation, and strong interferon response induction by positive feedback of immune response activation. Confirmation of the immune regulatory function of SLIRP in defense against autoimmune diseases Sjögren's syndrome, coronavirus, and encephalomyocarditis virus infection. >
Professor Yoosik Kim remarked, "Through this study, we have identified SLIRP as a crucial protein that drives immune amplification via mt-dsRNAs. Given its dual role in autoimmune diseases and viral infections, SLIRP presents a promising target for immune regulation therapies across various inflammatory disease contexts."
The study, with Ph.D. student Do-Young Ku (first author) and M.S. student Ye-Won Yang (second author) from the Department of Chemical and Biomolecular Engineering at KAIST as primary contributors, was published online in the journal Cell Reports on April 19, 2025.
※ Paper title: SLIRP amplifies antiviral signaling via positive feedback regulation and contributes to autoimmune diseases※ Main authors: Do-Young Ku (KAIST, first author), Ye-Won Yang (KAIST, second author), Seunghee Cha (University of Florida, corresponding author), Yoosik Kim (KAIST, corresponding author)
This study was supported by the Ministry of Health and Welfare's Public Health Technology Research Program and the National Institutes of Health (NIH) through Research Project (R01) funding.
2025.05.14 View 2186
KAIST Discovers Protein Switch that Turns Anti-Viral Immune Response On and Off
Even after the COVID-19 pandemic, various new infectious diseases continue to emerge, posing ongoing viral threats that demand robust and sustained immune defenses. However, excessive immune reactions can also harm body tissues, causing significant health issues. KAIST and an international research team have discovered a critical protein that acts as a 'switch' regulating immune responses to viruses. This breakthrough is expected to lay the groundwork for future infectious disease responses and autoimmune disease treatment strategies.
KAIST (President Kwang-Hyung Lee) announced on May 14 that a joint research team led by Professor Yoosik Kim from the Department of Chemical and Biomolecular Engineering at KAIST and Professor Seunghee Cha from University of Florida has discovered the mechanism by which double-stranded RNA derived from mitochondria amplifies immune responses. They identified the protein SLIRP as an 'immune switch' that regulates this process, playing a crucial role in both viral infections and autoimmune diseases.
< (From left) Master's candidate Yewon Yang, Professor Yoosik Kim and Ph.D. candidate Doyeong Ku of the Department of Chemical and Biomolecular Engineering >
Autoimmune diseases arise when the immune system fails to differentiate between external pathogens and the body's own molecules, leading to self-directed attacks. Despite extensive research, the precise causes of excessive inflammatory conditions like Sjögren’s syndrome and systemic lupus erythematosus remain unclear, and effective treatments are still limited.
To uncover the molecular mechanisms driving immune hyperactivation and to identify potential regulatory factors, the research team led by Professor Yoosik Kim focused on mitochondrial double-stranded RNA (mt-dsRNA), a genetic immunogenic material produced within cellular organelles. Since mt-dsRNA structurally resembles viral RNA, it can mistakenly trigger immune responses even in the absence of an actual viral infection.
The team discovered that SLIRP, a key regulator of mt-dsRNA, amplifies immune responses by stabilizing the RNA. They confirmed that SLIRP expression increases in experimental models simulating the tissues of autoimmune disease patients and viral infections. Conversely, suppressing SLIRP significantly reduced the immune response, underscoring its role as a critical factor in immune amplification.
This study also demonstrated the dual function of SLIRP in different contexts. In cells infected with human beta coronavirus OC43 and encephalomyocarditis virus (EMCV), SLIRP suppression led to reduced antiviral responses and increased viral replication. Meanwhile, in the blood and salivary gland cells of Sjögren’s syndrome patients, where both SLIRP and mt-dsRNA levels were elevated, suppressing SLIRP alleviated the abnormal immune response.
These findings highlight SLIRP as a key molecular switch that regulates immune responses in both infections and autoimmune diseases.
< Figure 1. Schematic diagram of antiviral signal amplification by SLIRP: SLIRP-based mt-dsRNA induction, cytoplasmic accumulation, and strong interferon response induction by positive feedback of immune response activation. Confirmation of the immune regulatory function of SLIRP in defense against autoimmune diseases Sjögren's syndrome, coronavirus, and encephalomyocarditis virus infection. >
Professor Yoosik Kim remarked, "Through this study, we have identified SLIRP as a crucial protein that drives immune amplification via mt-dsRNAs. Given its dual role in autoimmune diseases and viral infections, SLIRP presents a promising target for immune regulation therapies across various inflammatory disease contexts."
The study, with Ph.D. student Do-Young Ku (first author) and M.S. student Ye-Won Yang (second author) from the Department of Chemical and Biomolecular Engineering at KAIST as primary contributors, was published online in the journal Cell Reports on April 19, 2025.
※ Paper title: SLIRP amplifies antiviral signaling via positive feedback regulation and contributes to autoimmune diseases※ Main authors: Do-Young Ku (KAIST, first author), Ye-Won Yang (KAIST, second author), Seunghee Cha (University of Florida, corresponding author), Yoosik Kim (KAIST, corresponding author)
This study was supported by the Ministry of Health and Welfare's Public Health Technology Research Program and the National Institutes of Health (NIH) through Research Project (R01) funding.
2025.05.14 View 2186 -
 KAIST's Pioneering VR Precision Technology & Choreography Tool Receive Spotlights at CHI 2025
Accurate pointing in virtual spaces is essential for seamless interaction. If pointing is not precise, selecting the desired object becomes challenging, breaking user immersion and reducing overall experience quality. KAIST researchers have developed a technology that offers a vivid, lifelike experience in virtual space, alongside a new tool that assists choreographers throughout the creative process.
KAIST (President Kwang-Hyung Lee) announced on May 13th that a research team led by Professor Sang Ho Yoon of the Graduate School of Culture Technology, in collaboration with Professor Yang Zhang of the University of California, Los Angeles (UCLA), has developed the ‘T2IRay’ technology and the ‘ChoreoCraft’ platform, which enables choreographers to work more freely and creatively in virtual reality. These technologies received two Honorable Mention awards, recognizing the top 5% of papers, at CHI 2025*, the best international conference in the field of human-computer interaction, hosted by the Association for Computing Machinery (ACM) from April 25 to May 1.
< (From left) PhD candidates Jina Kim and Kyungeun Jung along with Master's candidate, Hyunyoung Han and Professor Sang Ho Yoon of KAIST Graduate School of Culture Technology and Professor Yang Zhang (top) of UCLA >
T2IRay: Enabling Virtual Input with Precision
T2IRay introduces a novel input method that allows for precise object pointing in virtual environments by expanding traditional thumb-to-index gestures. This approach overcomes previous limitations, such as interruptions or reduced accuracy due to changes in hand position or orientation.
The technology uses a local coordinate system based on finger relationships, ensuring continuous input even as hand positions shift. It accurately captures subtle thumb movements within this coordinate system, integrating natural head movements to allow fluid, intuitive control across a wide range.
< Figure 1. T2IRay framework utilizing the delicate movements of the thumb and index fingers for AR/VR pointing >
Professor Sang Ho Yoon explained, “T2IRay can significantly enhance the user experience in AR/VR by enabling smooth, stable control even when the user’s hands are in motion.”
This study, led by first author Jina Kim, was supported by the Excellent New Researcher Support Project of the National Research Foundation of Korea under the Ministry of Science and ICT, as well as the University ICT Research Center (ITRC) Support Project of the Institute of Information and Communications Technology Planning and Evaluation (IITP).
▴ Paper title: T2IRay: Design of Thumb-to-Index Based Indirect Pointing for Continuous and Robust AR/VR Input▴ Paper link: https://doi.org/10.1145/3706598.3713442
▴ T2IRay demo video: https://youtu.be/ElJlcJbkJPY
ChoreoCraft: Creativity Support through VR for Choreographers
In addition, Professor Yoon’s team developed ‘ChoreoCraft,’ a virtual reality tool designed to support choreographers by addressing the unique challenges they face, such as memorizing complex movements, overcoming creative blocks, and managing subjective feedback.
ChoreoCraft reduces reliance on memory by allowing choreographers to save and refine movements directly within a VR space, using a motion-capture avatar for real-time interaction. It also enhances creativity by suggesting movements that naturally fit with prior choreography and musical elements. Furthermore, the system provides quantitative feedback by analyzing kinematic factors like motion stability and engagement, helping choreographers make data-driven creative decisions.
< Figure 2. ChoreoCraft's approaches to encourage creative process >
Professor Yoon noted, “ChoreoCraft is a tool designed to address the core challenges faced by choreographers, enhancing both creativity and efficiency. In user tests with professional choreographers, it received high marks for its ability to spark creative ideas and provide valuable quantitative feedback.”
This research was conducted in collaboration with doctoral candidate Kyungeun Jung and master’s candidate Hyunyoung Han, alongside the Electronics and Telecommunications Research Institute (ETRI) and One Million Co., Ltd. (CEO Hye-rang Kim), with support from the Cultural and Arts Immersive Service Development Project by the Ministry of Culture, Sports and Tourism.
▴ Paper title: ChoreoCraft: In-situ Crafting of Choreography in Virtual Reality through Creativity Support Tools▴ Paper link: https://doi.org/10.1145/3706598.3714220
▴ ChoreoCraft demo video: https://youtu.be/Ms1fwiSBjjw
*CHI (Conference on Human Factors in Computing Systems): The premier international conference on human-computer interaction, organized by the ACM, was held this year from April 25 to May 1, 2025.
2025.05.13 View 2894
KAIST's Pioneering VR Precision Technology & Choreography Tool Receive Spotlights at CHI 2025
Accurate pointing in virtual spaces is essential for seamless interaction. If pointing is not precise, selecting the desired object becomes challenging, breaking user immersion and reducing overall experience quality. KAIST researchers have developed a technology that offers a vivid, lifelike experience in virtual space, alongside a new tool that assists choreographers throughout the creative process.
KAIST (President Kwang-Hyung Lee) announced on May 13th that a research team led by Professor Sang Ho Yoon of the Graduate School of Culture Technology, in collaboration with Professor Yang Zhang of the University of California, Los Angeles (UCLA), has developed the ‘T2IRay’ technology and the ‘ChoreoCraft’ platform, which enables choreographers to work more freely and creatively in virtual reality. These technologies received two Honorable Mention awards, recognizing the top 5% of papers, at CHI 2025*, the best international conference in the field of human-computer interaction, hosted by the Association for Computing Machinery (ACM) from April 25 to May 1.
< (From left) PhD candidates Jina Kim and Kyungeun Jung along with Master's candidate, Hyunyoung Han and Professor Sang Ho Yoon of KAIST Graduate School of Culture Technology and Professor Yang Zhang (top) of UCLA >
T2IRay: Enabling Virtual Input with Precision
T2IRay introduces a novel input method that allows for precise object pointing in virtual environments by expanding traditional thumb-to-index gestures. This approach overcomes previous limitations, such as interruptions or reduced accuracy due to changes in hand position or orientation.
The technology uses a local coordinate system based on finger relationships, ensuring continuous input even as hand positions shift. It accurately captures subtle thumb movements within this coordinate system, integrating natural head movements to allow fluid, intuitive control across a wide range.
< Figure 1. T2IRay framework utilizing the delicate movements of the thumb and index fingers for AR/VR pointing >
Professor Sang Ho Yoon explained, “T2IRay can significantly enhance the user experience in AR/VR by enabling smooth, stable control even when the user’s hands are in motion.”
This study, led by first author Jina Kim, was supported by the Excellent New Researcher Support Project of the National Research Foundation of Korea under the Ministry of Science and ICT, as well as the University ICT Research Center (ITRC) Support Project of the Institute of Information and Communications Technology Planning and Evaluation (IITP).
▴ Paper title: T2IRay: Design of Thumb-to-Index Based Indirect Pointing for Continuous and Robust AR/VR Input▴ Paper link: https://doi.org/10.1145/3706598.3713442
▴ T2IRay demo video: https://youtu.be/ElJlcJbkJPY
ChoreoCraft: Creativity Support through VR for Choreographers
In addition, Professor Yoon’s team developed ‘ChoreoCraft,’ a virtual reality tool designed to support choreographers by addressing the unique challenges they face, such as memorizing complex movements, overcoming creative blocks, and managing subjective feedback.
ChoreoCraft reduces reliance on memory by allowing choreographers to save and refine movements directly within a VR space, using a motion-capture avatar for real-time interaction. It also enhances creativity by suggesting movements that naturally fit with prior choreography and musical elements. Furthermore, the system provides quantitative feedback by analyzing kinematic factors like motion stability and engagement, helping choreographers make data-driven creative decisions.
< Figure 2. ChoreoCraft's approaches to encourage creative process >
Professor Yoon noted, “ChoreoCraft is a tool designed to address the core challenges faced by choreographers, enhancing both creativity and efficiency. In user tests with professional choreographers, it received high marks for its ability to spark creative ideas and provide valuable quantitative feedback.”
This research was conducted in collaboration with doctoral candidate Kyungeun Jung and master’s candidate Hyunyoung Han, alongside the Electronics and Telecommunications Research Institute (ETRI) and One Million Co., Ltd. (CEO Hye-rang Kim), with support from the Cultural and Arts Immersive Service Development Project by the Ministry of Culture, Sports and Tourism.
▴ Paper title: ChoreoCraft: In-situ Crafting of Choreography in Virtual Reality through Creativity Support Tools▴ Paper link: https://doi.org/10.1145/3706598.3714220
▴ ChoreoCraft demo video: https://youtu.be/Ms1fwiSBjjw
*CHI (Conference on Human Factors in Computing Systems): The premier international conference on human-computer interaction, organized by the ACM, was held this year from April 25 to May 1, 2025.
2025.05.13 View 2894 -
 KAIST Develops Novel Catalyst With 100-Fold Platinum Efficiency
Propylene, a key building block used in producing plastics, textiles, automotive components, and electronics, is essential to the petrochemical industry. A KAIST research team has developed a novel catalyst that dramatically enhances the efficiency of propylene production while significantly reducing costs.
< Photo. Professor Minkee Choi (left), and Ph.D. Candidate Susung Lee (right) of the Department of Chemical and Biomolecular Engineering >
KAIST (represented by President Kwang-Hyung Lee) announced on the 12th of May that a research group led by Professor Minkee Choi from the Department of Chemical and Biomolecular Engineering has successfully developed a new catalyst using inexpensive metals—gallium (Ga) and alumina (Al₂O₃)—with only a trace amount of platinum (100 ppm, or 0.01%). Remarkably, this new catalyst outperforms conventional industrial catalysts that use high concentrations of platinum (10,000 ppm).
Propylene is commonly produced through the propane dehydrogenation (PDH) process, which removes hydrogen from propane. Platinum has long been used as a catalyst in PDH due to its high efficiency in breaking carbon-hydrogen bonds and facilitating hydrogen removal. However, platinum is costly and suffers from performance degradation over repeated use.
To address this, the KAIST team engineered a catalyst that incorporates only a minimal amount of platinum, relying on gallium and alumina as the primary components.
< Figure 1. Schematic diagram showing the catalytic cooperation between gallium (Ga) and platinum (Pt) >
The core mechanism of the catalyst involves a cooperative function between the metals: gallium activates the C–H bond in propane to produce propylene, while platinum bonds the residual hydrogen atoms on the surface to form hydrogen gas (H₂), which is then released. This division of roles allows for high catalytic efficiency despite the drastic reduction in platinum content.
The researchers identified an optimal platinum-to-gallium ratio that delivered peak performance and provided a scientific rationale and quantitative metrics to predict this ideal composition.
Additionally, the team addressed a major limitation of traditional platinum catalysts: sintering—the agglomeration of platinum particles during repeated use, which causes performance loss. By adding a small amount of cerium (Ce), the researchers successfully suppressed this aggregation. As a result, the new catalyst maintained stable performance even after more than 20 reaction-regeneration cycles.
< Figure 2. Performance comparison of KAIST's newly developed catalyst (100 ppm platinum) and existing commercial platinum catalyst (10,000 ppm platinum) >
Professor Choi stated, “This research demonstrates the possibility of reducing platinum usage to 1/100th of current levels without compromising, and even enhancing, performance. It presents significant economic and environmental advantages, including lower catalyst costs, extended replacement intervals, and reduced catalyst waste.”
He added, “We are planning to evaluate this technology for large-scale process demonstration and commercialization. If adopted in industry, it could greatly improve the economic viability and efficiency of propylene production.”
The study was led by Professor Minkee Choi as corresponding author, with Ph.D. candidate Susung Lee as the first author. The findings were published in the Journal of the American Chemical Society (JACS), a leading journal in chemistry and chemical engineering, on February 13.※ Paper title: Ideal Bifunctional Catalysis for Propane Dehydrogenation over Pt-Promoted Gallia-Alumina and Minimized Use of Precious Elements※ DOI: https://pubs.acs.org/doi/10.1021/jacs.4c13787
The research was supported by the National Research Foundation of Korea and Hanwha Solutions Corporation.
2025.05.12 View 1317
KAIST Develops Novel Catalyst With 100-Fold Platinum Efficiency
Propylene, a key building block used in producing plastics, textiles, automotive components, and electronics, is essential to the petrochemical industry. A KAIST research team has developed a novel catalyst that dramatically enhances the efficiency of propylene production while significantly reducing costs.
< Photo. Professor Minkee Choi (left), and Ph.D. Candidate Susung Lee (right) of the Department of Chemical and Biomolecular Engineering >
KAIST (represented by President Kwang-Hyung Lee) announced on the 12th of May that a research group led by Professor Minkee Choi from the Department of Chemical and Biomolecular Engineering has successfully developed a new catalyst using inexpensive metals—gallium (Ga) and alumina (Al₂O₃)—with only a trace amount of platinum (100 ppm, or 0.01%). Remarkably, this new catalyst outperforms conventional industrial catalysts that use high concentrations of platinum (10,000 ppm).
Propylene is commonly produced through the propane dehydrogenation (PDH) process, which removes hydrogen from propane. Platinum has long been used as a catalyst in PDH due to its high efficiency in breaking carbon-hydrogen bonds and facilitating hydrogen removal. However, platinum is costly and suffers from performance degradation over repeated use.
To address this, the KAIST team engineered a catalyst that incorporates only a minimal amount of platinum, relying on gallium and alumina as the primary components.
< Figure 1. Schematic diagram showing the catalytic cooperation between gallium (Ga) and platinum (Pt) >
The core mechanism of the catalyst involves a cooperative function between the metals: gallium activates the C–H bond in propane to produce propylene, while platinum bonds the residual hydrogen atoms on the surface to form hydrogen gas (H₂), which is then released. This division of roles allows for high catalytic efficiency despite the drastic reduction in platinum content.
The researchers identified an optimal platinum-to-gallium ratio that delivered peak performance and provided a scientific rationale and quantitative metrics to predict this ideal composition.
Additionally, the team addressed a major limitation of traditional platinum catalysts: sintering—the agglomeration of platinum particles during repeated use, which causes performance loss. By adding a small amount of cerium (Ce), the researchers successfully suppressed this aggregation. As a result, the new catalyst maintained stable performance even after more than 20 reaction-regeneration cycles.
< Figure 2. Performance comparison of KAIST's newly developed catalyst (100 ppm platinum) and existing commercial platinum catalyst (10,000 ppm platinum) >
Professor Choi stated, “This research demonstrates the possibility of reducing platinum usage to 1/100th of current levels without compromising, and even enhancing, performance. It presents significant economic and environmental advantages, including lower catalyst costs, extended replacement intervals, and reduced catalyst waste.”
He added, “We are planning to evaluate this technology for large-scale process demonstration and commercialization. If adopted in industry, it could greatly improve the economic viability and efficiency of propylene production.”
The study was led by Professor Minkee Choi as corresponding author, with Ph.D. candidate Susung Lee as the first author. The findings were published in the Journal of the American Chemical Society (JACS), a leading journal in chemistry and chemical engineering, on February 13.※ Paper title: Ideal Bifunctional Catalysis for Propane Dehydrogenation over Pt-Promoted Gallia-Alumina and Minimized Use of Precious Elements※ DOI: https://pubs.acs.org/doi/10.1021/jacs.4c13787
The research was supported by the National Research Foundation of Korea and Hanwha Solutions Corporation.
2025.05.12 View 1317 -
 KAIST & CMU Unveils Amuse, a Songwriting AI-Collaborator to Help Create Music
Wouldn't it be great if music creators had someone to brainstorm with, help them when they're stuck, and explore different musical directions together? Researchers of KAIST and Carnegie Mellon University (CMU) have developed AI technology similar to a fellow songwriter who helps create music.
KAIST (President Kwang-Hyung Lee) has developed an AI-based music creation support system, Amuse, by a research team led by Professor Sung-Ju Lee of the School of Electrical Engineering in collaboration with CMU. The research was presented at the ACM Conference on Human Factors in Computing Systems (CHI), one of the world’s top conferences in human-computer interaction, held in Yokohama, Japan from April 26 to May 1. It received the Best Paper Award, given to only the top 1% of all submissions.
< (From left) Professor Chris Donahue of Carnegie Mellon University, Ph.D. Student Yewon Kim and Professor Sung-Ju Lee of the School of Electrical Engineering >
The system developed by Professor Sung-Ju Lee’s research team, Amuse, is an AI-based system that converts various forms of inspiration such as text, images, and audio into harmonic structures (chord progressions) to support composition.
For example, if a user inputs a phrase, image, or sound clip such as “memories of a warm summer beach”, Amuse automatically generates and suggests chord progressions that match the inspiration.
Unlike existing generative AI, Amuse is differentiated in that it respects the user's creative flow and naturally induces creative exploration through an interactive method that allows flexible integration and modification of AI suggestions.
The core technology of the Amuse system is a generation method that blends two approaches: a large language model creates music code based on the user's prompt and inspiration, while another AI model, trained on real music data, filters out awkward or unnatural results using rejection sampling.
< Figure 1. Amuse system configuration. After extracting music keywords from user input, a large language model-based code progression is generated and refined through rejection sampling (left). Code extraction from audio input is also possible (right). The bottom is an example visualizing the chord structure of the generated code. >
The research team conducted a user study targeting actual musicians and evaluated that Amuse has high potential as a creative companion, or a Co-Creative AI, a concept in which people and AI collaborate, rather than having a generative AI simply put together a song.
The paper, in which a Ph.D. student Yewon Kim and Professor Sung-Ju Lee of KAIST School of Electrical and Electronic Engineering and Carnegie Mellon University Professor Chris Donahue participated, demonstrated the potential of creative AI system design in both academia and industry. ※ Paper title: Amuse: Human-AI Collaborative Songwriting with Multimodal Inspirations DOI: https://doi.org/10.1145/3706598.3713818
※ Research demo video: https://youtu.be/udilkRSnftI?si=FNXccC9EjxHOCrm1
※ Research homepage: https://nmsl.kaist.ac.kr/projects/amuse/
Professor Sung-Ju Lee said, “Recent generative AI technology has raised concerns in that it directly imitates copyrighted content, thereby violating the copyright of the creator, or generating results one-way regardless of the creator’s intention. Accordingly, the research team was aware of this trend, paid attention to what the creator actually needs, and focused on designing an AI system centered on the creator.”
He continued, “Amuse is an attempt to explore the possibility of collaboration with AI while maintaining the initiative of the creator, and is expected to be a starting point for suggesting a more creator-friendly direction in the development of music creation tools and generative AI systems in the future.”
This research was conducted with the support of the National Research Foundation of Korea with funding from the government (Ministry of Science and ICT). (RS-2024-00337007)
2025.05.07 View 4282
KAIST & CMU Unveils Amuse, a Songwriting AI-Collaborator to Help Create Music
Wouldn't it be great if music creators had someone to brainstorm with, help them when they're stuck, and explore different musical directions together? Researchers of KAIST and Carnegie Mellon University (CMU) have developed AI technology similar to a fellow songwriter who helps create music.
KAIST (President Kwang-Hyung Lee) has developed an AI-based music creation support system, Amuse, by a research team led by Professor Sung-Ju Lee of the School of Electrical Engineering in collaboration with CMU. The research was presented at the ACM Conference on Human Factors in Computing Systems (CHI), one of the world’s top conferences in human-computer interaction, held in Yokohama, Japan from April 26 to May 1. It received the Best Paper Award, given to only the top 1% of all submissions.
< (From left) Professor Chris Donahue of Carnegie Mellon University, Ph.D. Student Yewon Kim and Professor Sung-Ju Lee of the School of Electrical Engineering >
The system developed by Professor Sung-Ju Lee’s research team, Amuse, is an AI-based system that converts various forms of inspiration such as text, images, and audio into harmonic structures (chord progressions) to support composition.
For example, if a user inputs a phrase, image, or sound clip such as “memories of a warm summer beach”, Amuse automatically generates and suggests chord progressions that match the inspiration.
Unlike existing generative AI, Amuse is differentiated in that it respects the user's creative flow and naturally induces creative exploration through an interactive method that allows flexible integration and modification of AI suggestions.
The core technology of the Amuse system is a generation method that blends two approaches: a large language model creates music code based on the user's prompt and inspiration, while another AI model, trained on real music data, filters out awkward or unnatural results using rejection sampling.
< Figure 1. Amuse system configuration. After extracting music keywords from user input, a large language model-based code progression is generated and refined through rejection sampling (left). Code extraction from audio input is also possible (right). The bottom is an example visualizing the chord structure of the generated code. >
The research team conducted a user study targeting actual musicians and evaluated that Amuse has high potential as a creative companion, or a Co-Creative AI, a concept in which people and AI collaborate, rather than having a generative AI simply put together a song.
The paper, in which a Ph.D. student Yewon Kim and Professor Sung-Ju Lee of KAIST School of Electrical and Electronic Engineering and Carnegie Mellon University Professor Chris Donahue participated, demonstrated the potential of creative AI system design in both academia and industry. ※ Paper title: Amuse: Human-AI Collaborative Songwriting with Multimodal Inspirations DOI: https://doi.org/10.1145/3706598.3713818
※ Research demo video: https://youtu.be/udilkRSnftI?si=FNXccC9EjxHOCrm1
※ Research homepage: https://nmsl.kaist.ac.kr/projects/amuse/
Professor Sung-Ju Lee said, “Recent generative AI technology has raised concerns in that it directly imitates copyrighted content, thereby violating the copyright of the creator, or generating results one-way regardless of the creator’s intention. Accordingly, the research team was aware of this trend, paid attention to what the creator actually needs, and focused on designing an AI system centered on the creator.”
He continued, “Amuse is an attempt to explore the possibility of collaboration with AI while maintaining the initiative of the creator, and is expected to be a starting point for suggesting a more creator-friendly direction in the development of music creation tools and generative AI systems in the future.”
This research was conducted with the support of the National Research Foundation of Korea with funding from the government (Ministry of Science and ICT). (RS-2024-00337007)
2025.05.07 View 4282 -
 Editing Parkinson's Disease – KAIST Makes World's First Discovery of an Inflammatory RNA Editing Enzyme through Co-work with UCL Researchers
< Professor Minee Choi of the Department of Brain and Cognitive Sciences (top left). Professor Sonia Gandhi (top right) and Professor Klenerman of the University College London (bottom right) >
Parkinson's disease (PD) is a neurodegenerative disorder in which the α-synuclein protein abnormally aggregates within brain cells, causing neuronal damage. Through international collaboration, researchers at KAIST have revealed that RNA editing plays a crucial role in regulating neuroinflammation, a key pathology of Parkinson's disease.
KAIST (represented by President Kwang-Hyung Lee) announced on the 27th of April that a research team led by Professor Minee L. Choi from the Department of Brain and Cognitive Sciences, in collaboration with University College London (UCL) and the Francis Crick Institute, discovered that the RNA editing enzyme ADAR1 plays an important role in controlling immune responses in astrocytes, glial cells that trigger protective reactions in the brain, and demonstrated that this mechanism is critically involved in the progression of Parkinson’s disease.
Professor Choi's research team created a co-culture model composed of astrocytes and neurons derived from stem cells originating from Parkinson's disease patients, in order to study the inflammatory responses of brain immune cells. They then treated the model with α-synuclein aggregates, which are known to cause Parkinson’s disease, and analyzed how the immune cells' inflammatory responses changed.
< Figure 1. Schematic diagram of the inflammatory RNA editing model in Parkinson's disease >
As a result, it was found that early pathological forms of α-synuclein, known as oligomers, activated the Toll-like receptor pathway, which acts as a danger sensor in astrocytes, as well as the interferon response pathway, an immune signaling network that combats viruses and pathogens. During this process, the RNA editing enzyme ADAR1 was expressed and transformed into an isoform with an altered protein structure and function.
Notably, the RNA editing activity of ADAR1, which normally functions to regulate immune responses during viral infections by converting adenosine (A) to inosine (I) through a process known as A-to-I RNA editing, was found to be abnormally focused on genes that cause inflammation rather than operating under normal conditions. This phenomenon was observed not only in the patient-derived neuron models but also in postmortem brain tissues from actual Parkinson’s disease patients.
< Figure 2. Experimental design and inflammatory response induction in astrocytes following treatment with α-synuclein oligomers (abnormally folded protein fragments) >
This directly proves that the dysregulation of RNA editing induces chronic inflammatory responses in astrocytes, ultimately leading to neuronal toxicity and pathological progression.
This study is significant in that it newly identified the regulation of RNA editing within astrocytes as a key mechanism behind neuroinflammatory responses. In particular, it suggests that ADAR1 could serve as a novel genetic target for the treatment of Parkinson’s disease.
It is also noteworthy that the study reflected actual pathological characteristics of patients by utilizing patient-specific induced pluripotent stem cell-based precision models for brain diseases.
Professor Minee L. Choi stated, “This study demonstrates that the regulator of inflammation caused by protein aggregation operates at the new layer of RNA editing, offering a completely different therapeutic strategy from existing approaches to Parkinson's disease treatment." She further emphasized, “RNA editing technology could become an important turning point in the development of therapeutics for neuroinflammation.”
< Figure 3. When treated with α-synuclein oligomers, the causative agent of Parkinson's disease, A-to-I RNA editing is induced to change genetic information by ADAR in patient-derived stem cell-differentiated glial cells, confirming that α-synuclein is likely to be associated with the progression of Parkinson's disease through RNA editing >
This study was published in Science Advances on April 11, with Professor Choi listed as a co-first author.
Paper Title: Astrocytic RNA editing regulates the host immune response to alpha-synuclein, Science Advances Vol.11, Issue 15. (DOI:10.1126/sciadv.adp8504)
Lead Authors: Karishma D’Sa (UCL, Co-First Author), Minee L. Choi (KAIST, Co-First Author), Mina Ryten (UCL, Corresponding Author), Sonia Gandhi (Francis Crick Institute, University of Cambridge, Corresponding Author)
This research was supported by the Brain Research Program and the Excellent Young Researcher Program of the National Research Foundation of Korea, as well as KAIST’s Daekyo Cognitive Enhancement Program.
2025.05.02 View 3460
Editing Parkinson's Disease – KAIST Makes World's First Discovery of an Inflammatory RNA Editing Enzyme through Co-work with UCL Researchers
< Professor Minee Choi of the Department of Brain and Cognitive Sciences (top left). Professor Sonia Gandhi (top right) and Professor Klenerman of the University College London (bottom right) >
Parkinson's disease (PD) is a neurodegenerative disorder in which the α-synuclein protein abnormally aggregates within brain cells, causing neuronal damage. Through international collaboration, researchers at KAIST have revealed that RNA editing plays a crucial role in regulating neuroinflammation, a key pathology of Parkinson's disease.
KAIST (represented by President Kwang-Hyung Lee) announced on the 27th of April that a research team led by Professor Minee L. Choi from the Department of Brain and Cognitive Sciences, in collaboration with University College London (UCL) and the Francis Crick Institute, discovered that the RNA editing enzyme ADAR1 plays an important role in controlling immune responses in astrocytes, glial cells that trigger protective reactions in the brain, and demonstrated that this mechanism is critically involved in the progression of Parkinson’s disease.
Professor Choi's research team created a co-culture model composed of astrocytes and neurons derived from stem cells originating from Parkinson's disease patients, in order to study the inflammatory responses of brain immune cells. They then treated the model with α-synuclein aggregates, which are known to cause Parkinson’s disease, and analyzed how the immune cells' inflammatory responses changed.
< Figure 1. Schematic diagram of the inflammatory RNA editing model in Parkinson's disease >
As a result, it was found that early pathological forms of α-synuclein, known as oligomers, activated the Toll-like receptor pathway, which acts as a danger sensor in astrocytes, as well as the interferon response pathway, an immune signaling network that combats viruses and pathogens. During this process, the RNA editing enzyme ADAR1 was expressed and transformed into an isoform with an altered protein structure and function.
Notably, the RNA editing activity of ADAR1, which normally functions to regulate immune responses during viral infections by converting adenosine (A) to inosine (I) through a process known as A-to-I RNA editing, was found to be abnormally focused on genes that cause inflammation rather than operating under normal conditions. This phenomenon was observed not only in the patient-derived neuron models but also in postmortem brain tissues from actual Parkinson’s disease patients.
< Figure 2. Experimental design and inflammatory response induction in astrocytes following treatment with α-synuclein oligomers (abnormally folded protein fragments) >
This directly proves that the dysregulation of RNA editing induces chronic inflammatory responses in astrocytes, ultimately leading to neuronal toxicity and pathological progression.
This study is significant in that it newly identified the regulation of RNA editing within astrocytes as a key mechanism behind neuroinflammatory responses. In particular, it suggests that ADAR1 could serve as a novel genetic target for the treatment of Parkinson’s disease.
It is also noteworthy that the study reflected actual pathological characteristics of patients by utilizing patient-specific induced pluripotent stem cell-based precision models for brain diseases.
Professor Minee L. Choi stated, “This study demonstrates that the regulator of inflammation caused by protein aggregation operates at the new layer of RNA editing, offering a completely different therapeutic strategy from existing approaches to Parkinson's disease treatment." She further emphasized, “RNA editing technology could become an important turning point in the development of therapeutics for neuroinflammation.”
< Figure 3. When treated with α-synuclein oligomers, the causative agent of Parkinson's disease, A-to-I RNA editing is induced to change genetic information by ADAR in patient-derived stem cell-differentiated glial cells, confirming that α-synuclein is likely to be associated with the progression of Parkinson's disease through RNA editing >
This study was published in Science Advances on April 11, with Professor Choi listed as a co-first author.
Paper Title: Astrocytic RNA editing regulates the host immune response to alpha-synuclein, Science Advances Vol.11, Issue 15. (DOI:10.1126/sciadv.adp8504)
Lead Authors: Karishma D’Sa (UCL, Co-First Author), Minee L. Choi (KAIST, Co-First Author), Mina Ryten (UCL, Corresponding Author), Sonia Gandhi (Francis Crick Institute, University of Cambridge, Corresponding Author)
This research was supported by the Brain Research Program and the Excellent Young Researcher Program of the National Research Foundation of Korea, as well as KAIST’s Daekyo Cognitive Enhancement Program.
2025.05.02 View 3460 -
 KAIST sends out Music and Bio-Signs of Professor Kwon Ji-yong, a.k.a. G-Dragon, into Space to Pulsate through Universe and Resonate among Stars
KAIST (President Kwang-Hyung Lee) announced on the 10th of April that it successfully promoted the world’s first ‘Space Sound Source Transmission Project’ based on media art at the KAIST Space Research Institute on April 9th through collaboration between Professor Jinjoon Lee of the Graduate School of Culture Technology, a world-renowned media artist, and the global K-Pop artist, G-Dragon.
This project was proposed as part of the ‘AI Entertech Research Center’ being promoted by KAIST and Galaxy Corporation. It is a project to transmit the message and sound of G-Dragon (real name, Kwon Ji-yong), a singer/song writer affiliated with Galaxy Corporation and a visiting professor in the Department of Mechanical Engineering at KAIST, to space for the first time in the world.
This is a convergence project that combines science, technology, art, and popular music, and is a new form of ‘space culture content’ experiment that connects KAIST’s cutting-edge space technology, Professor Jinjoon Lee’s media art work, and G-Dragon’s voice and sound source containing his latest digital single, "HOME SWEET HOME".
< Photo 1. Professor Jinjoon Lee's Open Your Eyes Project "Iris"'s imagery projected on the 13m space antenna at the Space Research Institute >
This collaboration was planned with the theme of ‘emotional signals that expand the inner universe of humans to the outer universe.’ The image of G-Dragon’s iris was augmented through AI as a window into soul symbolizing his uniqueness and identity, and the new song “Home Sweet Home” was combined as an audio message containing the vibration of that emotion.
This was actually transmitted into space using a next-generation small satellite developed by KAIST Space Research Institute, completing a symbolic performance in which an individual’s inner universe is transmitted to outer space.
Professor Jinjoon Lee’s cinematic media art work “Iris” was unveiled at the site. This work was screened in the world’s first projection mapping method* on KAIST Space Research Institute’s 13m space antenna. This video was created using generative artificial intelligence (AI) technology based on the image of G-Dragon's iris, and combined with sound using the data of the sounds of Emile Bell rings – the bell that holds a thousand years of history, it presented an emotional art experience that transcends time and space.
*Projection Mapping: A technology that projects light and images onto actual structures to create visual changes, and is a method of expression that artistically reinterprets space.
This work is one of the major research achievements of KAIST TX Lab and Professor Lee based on new media technology based on biometric data such as iris, heartbeat, and brain waves.
Professor Jinjoon Lee said, "The iris is a symbol that reflects inner emotions and identity, so much so that it is called the 'mirror of the soul,' and this work sought to express 'the infinite universe seen from the inside of humanity' through G-Dragon's gaze."
< Photo 2. (From left) Professor Jinjoon Lee of the Graduate School of Culture Technology and G-Dragon (Visiting Professor Kwon Ji-yong of the Department of Mechanical Engineering) >
He continued, "The universe is a realm of technology as well as a stage for imagination and emotion, and I look forward to an encounter with the unknown through a new attempt to speak of art in the language of science including AI and imagine science in the form of art." “G-Dragon’s voice and music have now begun their journey to space,” said Yong-ho Choi, Galaxy Corporation’s Chief Happiness Officer (CHO). “This project is an act of leaving music as a legacy for humanity, while also having an important meaning of attempting to communicate with space.” He added, “This is a pioneering step to introduce human culture to space, and it will remain as a monumental performance that opens a new chapter in the history of music comparable to the Beatles.”
Galaxy Corporation is leading the future entertainment technology industry through its collaboration with KAIST, and was recently selected as the only entertainment technology company in a private meeting with Microsoft CEO Nadella. In particular, it is promoting the globalization of AI entertainment technology, receiving praise as a “pioneer of imagination” for new forms of AI entertainment content, including the AI contents for the deceased.
< Photo 3. Photo of G-Dragon's Home Sweet Home being sent into the space via Professor Jinjoon Lee's Space Sound Source Transmission Project >
Through this project, KAIST Space Research Institute presented new possibilities for utilizing satellite technology, and showed a model for science to connect with society in a more popular way.
KAIST President Kwang-Hyung Lee said, “KAIST is a place that always supports new imaginations and challenges,” and added, “We will continue to strive to continue creative research that no one has ever thought of, like this project that combines science, technology, and art.”
In the meantime, Galaxy Corporation, the agency of G-Dragon’s Professor Kwon Ji-yong, is an AI entertainment company that presents a new paradigm based on IP, media, tech, and entertainment convergence technology.
2025.04.10 View 4188
KAIST sends out Music and Bio-Signs of Professor Kwon Ji-yong, a.k.a. G-Dragon, into Space to Pulsate through Universe and Resonate among Stars
KAIST (President Kwang-Hyung Lee) announced on the 10th of April that it successfully promoted the world’s first ‘Space Sound Source Transmission Project’ based on media art at the KAIST Space Research Institute on April 9th through collaboration between Professor Jinjoon Lee of the Graduate School of Culture Technology, a world-renowned media artist, and the global K-Pop artist, G-Dragon.
This project was proposed as part of the ‘AI Entertech Research Center’ being promoted by KAIST and Galaxy Corporation. It is a project to transmit the message and sound of G-Dragon (real name, Kwon Ji-yong), a singer/song writer affiliated with Galaxy Corporation and a visiting professor in the Department of Mechanical Engineering at KAIST, to space for the first time in the world.
This is a convergence project that combines science, technology, art, and popular music, and is a new form of ‘space culture content’ experiment that connects KAIST’s cutting-edge space technology, Professor Jinjoon Lee’s media art work, and G-Dragon’s voice and sound source containing his latest digital single, "HOME SWEET HOME".
< Photo 1. Professor Jinjoon Lee's Open Your Eyes Project "Iris"'s imagery projected on the 13m space antenna at the Space Research Institute >
This collaboration was planned with the theme of ‘emotional signals that expand the inner universe of humans to the outer universe.’ The image of G-Dragon’s iris was augmented through AI as a window into soul symbolizing his uniqueness and identity, and the new song “Home Sweet Home” was combined as an audio message containing the vibration of that emotion.
This was actually transmitted into space using a next-generation small satellite developed by KAIST Space Research Institute, completing a symbolic performance in which an individual’s inner universe is transmitted to outer space.
Professor Jinjoon Lee’s cinematic media art work “Iris” was unveiled at the site. This work was screened in the world’s first projection mapping method* on KAIST Space Research Institute’s 13m space antenna. This video was created using generative artificial intelligence (AI) technology based on the image of G-Dragon's iris, and combined with sound using the data of the sounds of Emile Bell rings – the bell that holds a thousand years of history, it presented an emotional art experience that transcends time and space.
*Projection Mapping: A technology that projects light and images onto actual structures to create visual changes, and is a method of expression that artistically reinterprets space.
This work is one of the major research achievements of KAIST TX Lab and Professor Lee based on new media technology based on biometric data such as iris, heartbeat, and brain waves.
Professor Jinjoon Lee said, "The iris is a symbol that reflects inner emotions and identity, so much so that it is called the 'mirror of the soul,' and this work sought to express 'the infinite universe seen from the inside of humanity' through G-Dragon's gaze."
< Photo 2. (From left) Professor Jinjoon Lee of the Graduate School of Culture Technology and G-Dragon (Visiting Professor Kwon Ji-yong of the Department of Mechanical Engineering) >
He continued, "The universe is a realm of technology as well as a stage for imagination and emotion, and I look forward to an encounter with the unknown through a new attempt to speak of art in the language of science including AI and imagine science in the form of art." “G-Dragon’s voice and music have now begun their journey to space,” said Yong-ho Choi, Galaxy Corporation’s Chief Happiness Officer (CHO). “This project is an act of leaving music as a legacy for humanity, while also having an important meaning of attempting to communicate with space.” He added, “This is a pioneering step to introduce human culture to space, and it will remain as a monumental performance that opens a new chapter in the history of music comparable to the Beatles.”
Galaxy Corporation is leading the future entertainment technology industry through its collaboration with KAIST, and was recently selected as the only entertainment technology company in a private meeting with Microsoft CEO Nadella. In particular, it is promoting the globalization of AI entertainment technology, receiving praise as a “pioneer of imagination” for new forms of AI entertainment content, including the AI contents for the deceased.
< Photo 3. Photo of G-Dragon's Home Sweet Home being sent into the space via Professor Jinjoon Lee's Space Sound Source Transmission Project >
Through this project, KAIST Space Research Institute presented new possibilities for utilizing satellite technology, and showed a model for science to connect with society in a more popular way.
KAIST President Kwang-Hyung Lee said, “KAIST is a place that always supports new imaginations and challenges,” and added, “We will continue to strive to continue creative research that no one has ever thought of, like this project that combines science, technology, and art.”
In the meantime, Galaxy Corporation, the agency of G-Dragon’s Professor Kwon Ji-yong, is an AI entertainment company that presents a new paradigm based on IP, media, tech, and entertainment convergence technology.
2025.04.10 View 4188 -
 KAIST Identifies Master Regulator Blocking Immunotherapy, Paving the Way for a New Lung Cancer Treatment
Immune checkpoint inhibitors, a class of immunotherapies that help immune cells attack cancer more effectively, have revolutionized cancer treatment. However, fewer than 20% of patients respond to these treatments, highlighting the urgent need for new strategies tailored to both responders and non-responders.
KAIST researchers have discovered that 'DEAD-box helicases 54 (DDX54)', a type of RNA-binding protein, is the master regulator that hinders the effectiveness of immunotherapy—opening a new path for lung cancer treatment. This breakthrough technology has been transferred to faculty startup BioRevert Inc., where it is currently being developed as a companion therapeutic and is expected to enter clinical trials by 2028.
< Photo 1. (From left) Researcher Jungeun Lee, Professor Kwang-Hyun Cho and Postdoctoral Researcher Jeong-Ryeol Gong of the Department of Bio and Brain Engineering at KAIST >
KAIST (represented by President Kwang-Hyung Lee) announced on April 8 that a research team led by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering had identified DDX54 as a critical factor that determines the immune evasion capacity of lung cancer cells. They demonstrated that suppressing DDX54 enhances immune cell infiltration into tumors and significantly improves the efficacy of immunotherapy.
Immunotherapy using anti-PD-1 or anti-PD-L1 antibodies is considered a powerful approach in cancer treatment. However, its low response rate limits the number of patients who actually benefit.
To identify likely responders, tumor mutational burden (TMB) has recently been approved by the FDA as a key biomarker for immunotherapy. Cancers with high mutation rates are thought to be more responsive to immune checkpoint inhibitors. However, even tumors with high TMB can display an “immune-desert” phenotype—where immune cell infiltration is severely limited—resulting in poor treatment responses.
< Figure 1. DDX54 was identified as the master regulator that induces resistance to immunotherapy by orchestrating suppression of immune cell infiltration through cancer tissues as lung cancer cells become immune-evasive >
Professor Kwang-Hyun Cho's research team compared transcriptome and genome data of lung cancer patients with immune evasion capabilities through gene regulatory network analysis (A) and discovered DDX54, a master regulator that induces resistance to immunotherapy (B-F).
This study is especially significant in that it successfully demonstrated that suppressing DDX54 in immune-desert lung tumors can overcome immunotherapy resistance and improve treatment outcomes.
The team used transcriptomic and genomic data from immune-evasive lung cancer patients and employed systems biology techniques to infer gene regulatory networks. Through this analysis, they identified DDX54 as a central regulator in the immune evasion of lung cancer cells.
In a syngeneic mouse model, the suppression of DDX54 led to significant increases in the infiltration of anti-cancer immune cells such as T cells and NK cells, and greatly improved the response to immunotherapy.
Single-cell transcriptomic and spatial transcriptomic analyses further showed that combination therapy targeting DDX54 promoted the differentiation of T cells and memory T cells that suppress tumors, while reducing the infiltration of regulatory T cells and exhausted T cells that support tumor growth.
< Figure 2. In the syngeneic mouse model made of lung cancer cells, it was confirmed that inhibiting DDX54 reversed the immune-evasion ability of cancer cells and enhanced the sensitivity to anti-PD-1 therapy >
In a syngeneic mouse model made of lung cancer cells exhibiting immunotherapy resistance, the treatment applied after DDX54 inhibition resulted in statistically significant inhibition of lung cancer growth (B-D) and a significant increase in immune cell infiltration into the tumor tissue (E, F).
The mechanism is believed to involve DDX54 suppression inactivating signaling pathways such as JAK-STAT, MYC, and NF-κB, thereby downregulating immune-evasive proteins CD38 and CD47. This also reduced the infiltration of circulating monocytes—which promote tumor development—and promoted the differentiation of M1 macrophages that play anti-tumor roles.
Professor Kwang-Hyun Cho stated, “We have, for the first time, identified a master regulatory factor that enables immune evasion in lung cancer cells. By targeting this factor, we developed a new therapeutic strategy that can induce responsiveness to immunotherapy in previously resistant cancers.”
He added, “The discovery of DDX54—hidden within the complex molecular networks of cancer cells—was made possible through the systematic integration of systems biology, combining IT and BT.”
The study, led by Professor Kwang-Hyun Cho, was published in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on April 2, 2025, with Jeong-Ryeol Gong being the first author, Jungeun Lee, a co-first author, and Younghyun Han, a co-author of the article.
< Figure 3. Single-cell transcriptome and spatial transcriptome analysis confirmed that knockdown of DDX54 increased immune cell infiltration into cancer tissues >
In a syngeneic mouse model made of lung cancer cells that underwent immunotherapy in combination with DDX54 inhibition, single-cell transcriptome (H-L) and spatial transcriptome (A-G) analysis of immune cells infiltrating inside cancer tissues were performed. As a result, it was confirmed that anticancer immune cells such as T cells, B cells, and NK cells actively infiltrated the core of lung cancer tissues when DDX54 inhibition and immunotherapy were concurrently administered.
(Paper title: “DDX54 downregulation enhances anti-PD1 therapy in immune-desert lung tumors with high tumor mutational burden,” DOI: https://doi.org/10.1073/pnas.2412310122)
This work was supported by the Ministry of Science and ICT and the National Research Foundation of Korea through the Mid-Career Research Program and Basic Research Laboratory Program.
< Figure 4. The identified master regulator DDX54 was confirmed to induce CD38 and CD47 expression through Jak-Stat3, MYC, and NF-κB activation. >
DDX54 activates the Jak-Stat3, MYC, and NF-κB pathways in lung cancer cells to increase CD38 and CD47 expression (A-G). This creates a cancer microenvironment that contributes to cancer development (H) and ultimately induces immune anticancer treatment resistance.
< Figure 5. It was confirmed that an immune-inflamed environment can be created by combining DDX54 inhibition and immune checkpoint inhibitor (ICI) therapy. >
When DDX54 inhibition and ICI therapy are simultaneously administered, the cancer cell characteristics change, the immune evasion ability is restored, and the environment is transformed into an ‘immune-activated’ environment in which immune cells easily infiltrate cancer tissues. This strengthens the anticancer immune response, thereby increasing the sensitivity of immunotherapy even in lung cancer tissues that previously had low responsiveness to immunotherapy.
2025.04.08 View 4601
KAIST Identifies Master Regulator Blocking Immunotherapy, Paving the Way for a New Lung Cancer Treatment
Immune checkpoint inhibitors, a class of immunotherapies that help immune cells attack cancer more effectively, have revolutionized cancer treatment. However, fewer than 20% of patients respond to these treatments, highlighting the urgent need for new strategies tailored to both responders and non-responders.
KAIST researchers have discovered that 'DEAD-box helicases 54 (DDX54)', a type of RNA-binding protein, is the master regulator that hinders the effectiveness of immunotherapy—opening a new path for lung cancer treatment. This breakthrough technology has been transferred to faculty startup BioRevert Inc., where it is currently being developed as a companion therapeutic and is expected to enter clinical trials by 2028.
< Photo 1. (From left) Researcher Jungeun Lee, Professor Kwang-Hyun Cho and Postdoctoral Researcher Jeong-Ryeol Gong of the Department of Bio and Brain Engineering at KAIST >
KAIST (represented by President Kwang-Hyung Lee) announced on April 8 that a research team led by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering had identified DDX54 as a critical factor that determines the immune evasion capacity of lung cancer cells. They demonstrated that suppressing DDX54 enhances immune cell infiltration into tumors and significantly improves the efficacy of immunotherapy.
Immunotherapy using anti-PD-1 or anti-PD-L1 antibodies is considered a powerful approach in cancer treatment. However, its low response rate limits the number of patients who actually benefit.
To identify likely responders, tumor mutational burden (TMB) has recently been approved by the FDA as a key biomarker for immunotherapy. Cancers with high mutation rates are thought to be more responsive to immune checkpoint inhibitors. However, even tumors with high TMB can display an “immune-desert” phenotype—where immune cell infiltration is severely limited—resulting in poor treatment responses.
< Figure 1. DDX54 was identified as the master regulator that induces resistance to immunotherapy by orchestrating suppression of immune cell infiltration through cancer tissues as lung cancer cells become immune-evasive >
Professor Kwang-Hyun Cho's research team compared transcriptome and genome data of lung cancer patients with immune evasion capabilities through gene regulatory network analysis (A) and discovered DDX54, a master regulator that induces resistance to immunotherapy (B-F).
This study is especially significant in that it successfully demonstrated that suppressing DDX54 in immune-desert lung tumors can overcome immunotherapy resistance and improve treatment outcomes.
The team used transcriptomic and genomic data from immune-evasive lung cancer patients and employed systems biology techniques to infer gene regulatory networks. Through this analysis, they identified DDX54 as a central regulator in the immune evasion of lung cancer cells.
In a syngeneic mouse model, the suppression of DDX54 led to significant increases in the infiltration of anti-cancer immune cells such as T cells and NK cells, and greatly improved the response to immunotherapy.
Single-cell transcriptomic and spatial transcriptomic analyses further showed that combination therapy targeting DDX54 promoted the differentiation of T cells and memory T cells that suppress tumors, while reducing the infiltration of regulatory T cells and exhausted T cells that support tumor growth.
< Figure 2. In the syngeneic mouse model made of lung cancer cells, it was confirmed that inhibiting DDX54 reversed the immune-evasion ability of cancer cells and enhanced the sensitivity to anti-PD-1 therapy >
In a syngeneic mouse model made of lung cancer cells exhibiting immunotherapy resistance, the treatment applied after DDX54 inhibition resulted in statistically significant inhibition of lung cancer growth (B-D) and a significant increase in immune cell infiltration into the tumor tissue (E, F).
The mechanism is believed to involve DDX54 suppression inactivating signaling pathways such as JAK-STAT, MYC, and NF-κB, thereby downregulating immune-evasive proteins CD38 and CD47. This also reduced the infiltration of circulating monocytes—which promote tumor development—and promoted the differentiation of M1 macrophages that play anti-tumor roles.
Professor Kwang-Hyun Cho stated, “We have, for the first time, identified a master regulatory factor that enables immune evasion in lung cancer cells. By targeting this factor, we developed a new therapeutic strategy that can induce responsiveness to immunotherapy in previously resistant cancers.”
He added, “The discovery of DDX54—hidden within the complex molecular networks of cancer cells—was made possible through the systematic integration of systems biology, combining IT and BT.”
The study, led by Professor Kwang-Hyun Cho, was published in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on April 2, 2025, with Jeong-Ryeol Gong being the first author, Jungeun Lee, a co-first author, and Younghyun Han, a co-author of the article.
< Figure 3. Single-cell transcriptome and spatial transcriptome analysis confirmed that knockdown of DDX54 increased immune cell infiltration into cancer tissues >
In a syngeneic mouse model made of lung cancer cells that underwent immunotherapy in combination with DDX54 inhibition, single-cell transcriptome (H-L) and spatial transcriptome (A-G) analysis of immune cells infiltrating inside cancer tissues were performed. As a result, it was confirmed that anticancer immune cells such as T cells, B cells, and NK cells actively infiltrated the core of lung cancer tissues when DDX54 inhibition and immunotherapy were concurrently administered.
(Paper title: “DDX54 downregulation enhances anti-PD1 therapy in immune-desert lung tumors with high tumor mutational burden,” DOI: https://doi.org/10.1073/pnas.2412310122)
This work was supported by the Ministry of Science and ICT and the National Research Foundation of Korea through the Mid-Career Research Program and Basic Research Laboratory Program.
< Figure 4. The identified master regulator DDX54 was confirmed to induce CD38 and CD47 expression through Jak-Stat3, MYC, and NF-κB activation. >
DDX54 activates the Jak-Stat3, MYC, and NF-κB pathways in lung cancer cells to increase CD38 and CD47 expression (A-G). This creates a cancer microenvironment that contributes to cancer development (H) and ultimately induces immune anticancer treatment resistance.
< Figure 5. It was confirmed that an immune-inflamed environment can be created by combining DDX54 inhibition and immune checkpoint inhibitor (ICI) therapy. >
When DDX54 inhibition and ICI therapy are simultaneously administered, the cancer cell characteristics change, the immune evasion ability is restored, and the environment is transformed into an ‘immune-activated’ environment in which immune cells easily infiltrate cancer tissues. This strengthens the anticancer immune response, thereby increasing the sensitivity of immunotherapy even in lung cancer tissues that previously had low responsiveness to immunotherapy.
2025.04.08 View 4601 -
 KAIST Accelerates Synthetic Microbe Design by Discovering Novel Enzymes Using AI
< (From left) Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering (top), Hongkeun Ji, PhD candidate of the Department of Chemical and Biomolecular Engineering (top), Ha Rim Kim, PhD candidate of the Department of Chemical and Biomolecular Engineering, and Dr. Gi Bae Kim of the BioProcess Engineering Research Center >
Enzymes are proteins that catalyze biochemical reactions within cells and play a pivotal role in metabolic processes. Accordingly, identifying the functions of novel enzymes is a critical task in the construction of microbial cell factories.
A KAIST research team has leveraged artificial intelligence (AI) to design novel enzymes that do not exist in nature, significantly accelerating microbial cell factory development and boosting the potential for next-generation biotechnological applications such as drug development and biofuel production.
KAIST (represented by President Kwang-Hyung Lee) announced on the 21st of April that Distinguished Professor Sang Yup Lee and his team from the Department of Chemical and Biomolecular Engineering have published a review titled “Enzyme Functional Classification Using Artificial Intelligence,” which outlines the advancement of AI-based enzyme function prediction technologies and analyzes how AI has contributed to the discovery and design of new enzymes.
Professor Lee’s team systematically reviewed the development of enzyme function prediction technologies utilizing machine learning and deep learning, offering a comprehensive analysis.
From sequence similarity-based prediction methods to the integration of convolutional neural networks (CNNs), recurrent neural networks (RNNs), graph neural networks (GNNs), and transformer-based large language models, the paper covers a broad range of AI applications. It analyzes how these technologies extract meaningful information from protein sequences and enhance prediction accuracy.
In particular, enzyme function prediction using deep learning goes beyond simple sequence similarity analysis. By automatically extracting structural and evolutionary features embedded in amino acid sequences, deep learning enables more precise predictions of catalytic functions.
This highlights the unique advantages of AI models compared to traditional bioinformatics approaches.
Moreover, the review suggests that the advancement of generative AI will move future research beyond predicting existing functions to generating entirely new enzymes with functions not found in nature. This shift is expected to profoundly impact the trajectory of biotechnology and synthetic biology.
< Figure 1. Extraction of enzyme characteristics and function prediction using various deep learning structures >
Ha Rim Kim, a Ph.D. candidate and co-first author from the Department of Chemical and Biomolecular Engineering, stated, “AI-based enzyme function prediction and enzyme design are highly important across various fields including metabolic engineering, synthetic biology, and healthcare.”
Distinguished Professor Sang Yup Lee added, “AI-powered enzyme function prediction shows the potential to solve diverse biological problems and will significantly contribute to accelerating research across the entire field.”
The review was published on March 28 in Trends in Biotechnology, a leading biotechnology journal issued by Cell Press.
※ Title: Enzyme Functional Classification Using Artificial Intelligence
※DOI: https://doi.org/10.1016/j.tibtech.2025.03.003
※ Author Information: Ha Rim Kim (KAIST, Co-first author), Hongkeun Ji (KAIST, Co-first author), Gi Bae Kim (KAIST, Third author), Sang Yup Lee (KAIST, Corresponding author)
This research was supported by the Ministry of Science and ICT under the project Development of Core Technologies for Advanced Synthetic Biology to Lead the Bio-Manufacturing Industry (aimed at replacing petroleum-based chemicals), and also by joint support from the Ministry of Science and ICT and the Ministry of Health and Welfare for the project Development of Novel Antibiotic Structures Using Deep Learning-Based Synthetic Biology.
2025.04.07 View 2879
KAIST Accelerates Synthetic Microbe Design by Discovering Novel Enzymes Using AI
< (From left) Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering (top), Hongkeun Ji, PhD candidate of the Department of Chemical and Biomolecular Engineering (top), Ha Rim Kim, PhD candidate of the Department of Chemical and Biomolecular Engineering, and Dr. Gi Bae Kim of the BioProcess Engineering Research Center >
Enzymes are proteins that catalyze biochemical reactions within cells and play a pivotal role in metabolic processes. Accordingly, identifying the functions of novel enzymes is a critical task in the construction of microbial cell factories.
A KAIST research team has leveraged artificial intelligence (AI) to design novel enzymes that do not exist in nature, significantly accelerating microbial cell factory development and boosting the potential for next-generation biotechnological applications such as drug development and biofuel production.
KAIST (represented by President Kwang-Hyung Lee) announced on the 21st of April that Distinguished Professor Sang Yup Lee and his team from the Department of Chemical and Biomolecular Engineering have published a review titled “Enzyme Functional Classification Using Artificial Intelligence,” which outlines the advancement of AI-based enzyme function prediction technologies and analyzes how AI has contributed to the discovery and design of new enzymes.
Professor Lee’s team systematically reviewed the development of enzyme function prediction technologies utilizing machine learning and deep learning, offering a comprehensive analysis.
From sequence similarity-based prediction methods to the integration of convolutional neural networks (CNNs), recurrent neural networks (RNNs), graph neural networks (GNNs), and transformer-based large language models, the paper covers a broad range of AI applications. It analyzes how these technologies extract meaningful information from protein sequences and enhance prediction accuracy.
In particular, enzyme function prediction using deep learning goes beyond simple sequence similarity analysis. By automatically extracting structural and evolutionary features embedded in amino acid sequences, deep learning enables more precise predictions of catalytic functions.
This highlights the unique advantages of AI models compared to traditional bioinformatics approaches.
Moreover, the review suggests that the advancement of generative AI will move future research beyond predicting existing functions to generating entirely new enzymes with functions not found in nature. This shift is expected to profoundly impact the trajectory of biotechnology and synthetic biology.
< Figure 1. Extraction of enzyme characteristics and function prediction using various deep learning structures >
Ha Rim Kim, a Ph.D. candidate and co-first author from the Department of Chemical and Biomolecular Engineering, stated, “AI-based enzyme function prediction and enzyme design are highly important across various fields including metabolic engineering, synthetic biology, and healthcare.”
Distinguished Professor Sang Yup Lee added, “AI-powered enzyme function prediction shows the potential to solve diverse biological problems and will significantly contribute to accelerating research across the entire field.”
The review was published on March 28 in Trends in Biotechnology, a leading biotechnology journal issued by Cell Press.
※ Title: Enzyme Functional Classification Using Artificial Intelligence
※DOI: https://doi.org/10.1016/j.tibtech.2025.03.003
※ Author Information: Ha Rim Kim (KAIST, Co-first author), Hongkeun Ji (KAIST, Co-first author), Gi Bae Kim (KAIST, Third author), Sang Yup Lee (KAIST, Corresponding author)
This research was supported by the Ministry of Science and ICT under the project Development of Core Technologies for Advanced Synthetic Biology to Lead the Bio-Manufacturing Industry (aimed at replacing petroleum-based chemicals), and also by joint support from the Ministry of Science and ICT and the Ministry of Health and Welfare for the project Development of Novel Antibiotic Structures Using Deep Learning-Based Synthetic Biology.
2025.04.07 View 2879 -
 KAIST Develops Retinal Therapy to Restore Lost Vision
Vision is one of the most crucial human senses, yet over 300 million people worldwide are at risk of vision loss due to various retinal diseases. While recent advancements in retinal disease treatments have successfully slowed disease progression, no effective therapy has been developed to restore already lost vision—until now. KAIST researchers have successfully developed a novel drug to restore vision.
< Photo 1. (From left) Ph.D. candidate Museong Kim, Professor Jin Woo Kim, and Dr. Eun Jung Lee of KAIST Department of Biological Sciences >
KAIST (represented by President Kwang Hyung Lee) announced on the 30th of March that a research team led by Professor Jin Woo Kim from the Department of Biological Sciences has developed a treatment method that restores vision through retinal nerve regeneration.
The research team successfully induced retinal regeneration and vision recovery in a disease-model mouse by administering a compound that blocks the PROX1 (prospero homeobox 1) protein, which suppresses retinal regeneration. Furthermore, the effect lasted for more than six months.
This study marks the first successful induction of long-term neural regeneration in mammalian retinas, offering new hope to patients with degenerative retinal diseases who previously had no treatment options.
As the global population continues to age, the number of retinal disease patients is steadily increasing. However, no treatments exist to restore damaged retinas and vision. The primary reason for this is the mammalian retina's inability to regenerate once damaged.
Studies on cold-blooded animals, such as fish—known for their robust retinal regeneration—have shown that retinal injuries trigger Müller glia cells to dedifferentiate into retinal progenitor cells, which then generate new neurons. However, in mammals, this process is impaired, leading to permanent retinal damage.
< Figure 1. Schematic diagram of the mechanism of retinal regeneration through inhibition of PROX1 migration. PROX1 protein secreted from retinal damaged retinal neurons transfers to Müllerglia and inhibits dedifferentiation into neural progenitor cells and neural regeneration. When PROX1 is captured outside the cells by an antibody against PROX1 and its transfer to Müllerglia is interfered, dedifferentiation of Müllerglia cells and retinal regeneration processes are resumed, restoring visual function. >
Through this study, the research team identified the PROX1 protein as a key inhibitor of Müller glia dedifferentiation in mammals. PROX1 is a protein found in neurons of the retina, hippocampus, and spinal cord, where it suppresses neural stem cell proliferation and promotes differentiation into neurons.
The researchers discovered that PROX1 accumulates in damaged mouse retinal Müller glia, but is absent in the highly regenerative Müller glia of fish. Furthermore, they demonstrated that the PROX1 found in Müller glia is not synthesized internally but rather taken up from surrounding neurons, which fail to degrade and instead secrete the protein.
Based on this finding, the team developed a method to restore Müller glia’s regenerative ability by eliminating extracellular PROX1 before it reaches these cells.
< Figure 2. Retinal regeneration and visual recovery in a retinitis pigmentosa model mouse through Anti-PROX1 gene therapy. After administration of adeno-associated virus expressing PROX1 neutralizing antibodies (AAV2-Anti-PROX1) to the eyes of RP1 retinitis pigmentosa model mice with vision loss, the photoreceptor cell layer of the retina is restored (A) and vision is restored (B). >
This approach involves using an antibody that binds to PROX1, developed by Celliaz Inc., a biotech startup founded by Professor Jin Woo Kim’s research lab. When administered to disease-model mouse retinas, this antibody significantly promoted neural regeneration. Additionally, when delivered, the antibody gene to the retinas of retinitis pigmentosa disease model mice, it enabled sustained retinal regeneration and vision restoration for over six months.
The retinal regeneration-inducing therapy is currently being developed by Celliaz Inc. for application in various degenerative retinal diseases that currently lack effective treatments. The company aims to begin clinical trials by 2028.
This study was co-authored by Dr. Eun Jung Lee of Celliaz Inc. and Museong Kim, a Ph.D. candidate at KAIST, as joint first authors. The findings were published online on March 26 in the international journal Nature Communications. (Paper Title: Restoration of retinal regenerative potential of Müller glia by disrupting intercellular Prox1 transfer | DOI: 10.1038/s41467-025-58290-8)
Dr. Eun Jung Lee stated, "We are about completing the optimization of the PROX1-neutralizing antibody (CLZ001) and move to preclinical studies before administering it to retinal disease patients. Our goal is to provide a solution for patients at risk of blindness who currently lack proper treatment options."
This research was supported by research funds from Korean National Research Foundation (NRF) and the Korea Drug Development Foundation (KDDF).
2025.03.31 View 12315
KAIST Develops Retinal Therapy to Restore Lost Vision
Vision is one of the most crucial human senses, yet over 300 million people worldwide are at risk of vision loss due to various retinal diseases. While recent advancements in retinal disease treatments have successfully slowed disease progression, no effective therapy has been developed to restore already lost vision—until now. KAIST researchers have successfully developed a novel drug to restore vision.
< Photo 1. (From left) Ph.D. candidate Museong Kim, Professor Jin Woo Kim, and Dr. Eun Jung Lee of KAIST Department of Biological Sciences >
KAIST (represented by President Kwang Hyung Lee) announced on the 30th of March that a research team led by Professor Jin Woo Kim from the Department of Biological Sciences has developed a treatment method that restores vision through retinal nerve regeneration.
The research team successfully induced retinal regeneration and vision recovery in a disease-model mouse by administering a compound that blocks the PROX1 (prospero homeobox 1) protein, which suppresses retinal regeneration. Furthermore, the effect lasted for more than six months.
This study marks the first successful induction of long-term neural regeneration in mammalian retinas, offering new hope to patients with degenerative retinal diseases who previously had no treatment options.
As the global population continues to age, the number of retinal disease patients is steadily increasing. However, no treatments exist to restore damaged retinas and vision. The primary reason for this is the mammalian retina's inability to regenerate once damaged.
Studies on cold-blooded animals, such as fish—known for their robust retinal regeneration—have shown that retinal injuries trigger Müller glia cells to dedifferentiate into retinal progenitor cells, which then generate new neurons. However, in mammals, this process is impaired, leading to permanent retinal damage.
< Figure 1. Schematic diagram of the mechanism of retinal regeneration through inhibition of PROX1 migration. PROX1 protein secreted from retinal damaged retinal neurons transfers to Müllerglia and inhibits dedifferentiation into neural progenitor cells and neural regeneration. When PROX1 is captured outside the cells by an antibody against PROX1 and its transfer to Müllerglia is interfered, dedifferentiation of Müllerglia cells and retinal regeneration processes are resumed, restoring visual function. >
Through this study, the research team identified the PROX1 protein as a key inhibitor of Müller glia dedifferentiation in mammals. PROX1 is a protein found in neurons of the retina, hippocampus, and spinal cord, where it suppresses neural stem cell proliferation and promotes differentiation into neurons.
The researchers discovered that PROX1 accumulates in damaged mouse retinal Müller glia, but is absent in the highly regenerative Müller glia of fish. Furthermore, they demonstrated that the PROX1 found in Müller glia is not synthesized internally but rather taken up from surrounding neurons, which fail to degrade and instead secrete the protein.
Based on this finding, the team developed a method to restore Müller glia’s regenerative ability by eliminating extracellular PROX1 before it reaches these cells.
< Figure 2. Retinal regeneration and visual recovery in a retinitis pigmentosa model mouse through Anti-PROX1 gene therapy. After administration of adeno-associated virus expressing PROX1 neutralizing antibodies (AAV2-Anti-PROX1) to the eyes of RP1 retinitis pigmentosa model mice with vision loss, the photoreceptor cell layer of the retina is restored (A) and vision is restored (B). >
This approach involves using an antibody that binds to PROX1, developed by Celliaz Inc., a biotech startup founded by Professor Jin Woo Kim’s research lab. When administered to disease-model mouse retinas, this antibody significantly promoted neural regeneration. Additionally, when delivered, the antibody gene to the retinas of retinitis pigmentosa disease model mice, it enabled sustained retinal regeneration and vision restoration for over six months.
The retinal regeneration-inducing therapy is currently being developed by Celliaz Inc. for application in various degenerative retinal diseases that currently lack effective treatments. The company aims to begin clinical trials by 2028.
This study was co-authored by Dr. Eun Jung Lee of Celliaz Inc. and Museong Kim, a Ph.D. candidate at KAIST, as joint first authors. The findings were published online on March 26 in the international journal Nature Communications. (Paper Title: Restoration of retinal regenerative potential of Müller glia by disrupting intercellular Prox1 transfer | DOI: 10.1038/s41467-025-58290-8)
Dr. Eun Jung Lee stated, "We are about completing the optimization of the PROX1-neutralizing antibody (CLZ001) and move to preclinical studies before administering it to retinal disease patients. Our goal is to provide a solution for patients at risk of blindness who currently lack proper treatment options."
This research was supported by research funds from Korean National Research Foundation (NRF) and the Korea Drug Development Foundation (KDDF).
2025.03.31 View 12315 -
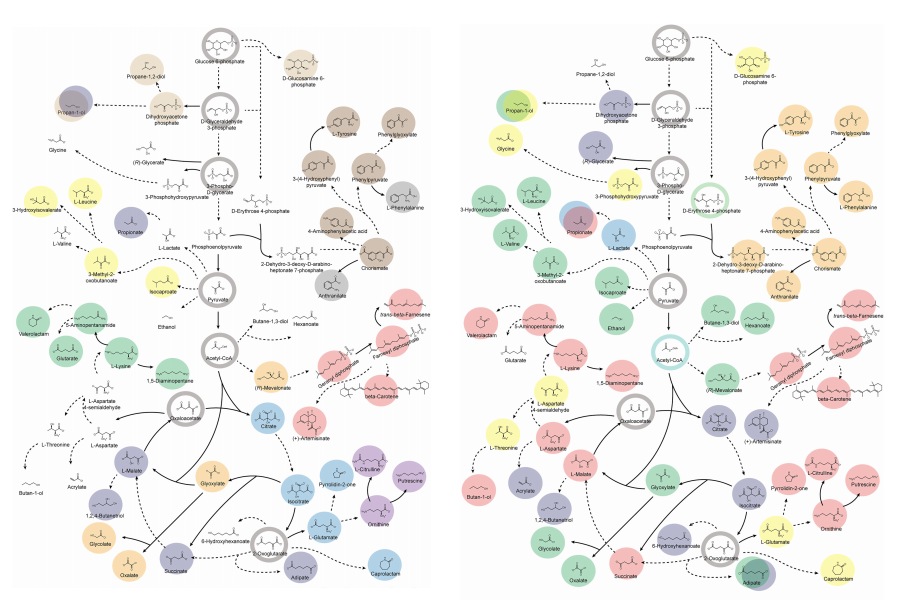 KAIST provides a comprehensive resource on microbial cell factories for sustainable chemical production
In silico analysis of five industrial microorganisms identifies optimal strains and metabolic engineering strategies for producing 235 valuable chemicals
Climate change and the depletion of fossil fuels have raised the global need for sustainable chemical production. In response to these environmental challenges, microbial cell factories are gaining attention as eco-friendly platforms for producing chemicals using renewable resources, while metabolic engineering technologies to enhance these cell factories are becoming crucial tools for maximizing production efficiency. However, difficulties in selecting suitable microbial strains and optimizing complex metabolic pathways continue to pose significant obstacles to practical industrial applications.
KAIST (President Kwang-Hyung Lee) announced on 27th of March that Distinguished Professor Sang Yup Lee’s research team in the Department of Chemical and Biomolecular Engineering comprehensively evaluated the production capabilities of various industrial microbial cell factories using in silico simulations and, based on these findings, identified the most suitable microbial strains for producing specific chemicals as well as optimal metabolic engineering strategies.
Previously, researchers attempted to determine the best strains and efficient metabolic engineering strategies among numerous microbial candidates through extensive biological experiments and meticulous verification processes. However, this approach required substantial time and costs. Recently, the introduction of genome-scale metabolic models (GEMs), which reconstruct the metabolic networks within an organism based on its entire genome information, has enabled systematic analysis of metabolic fluxes via computer simulations. This development offers a new way to overcome limitations of conventional experimental approaches, revolutionizing both strain selection and metabolic pathway design.
Accordingly, Professor Lee’s team at the Department of Chemical and Biomolecular Engineering, KAIST, evaluated the production capabilities of five representative industrial microorganisms—Escherichia coli, Saccharomyces cerevisiae, Bacillus subtilis, Corynebacterium glutamicum, and Pseudomonas putida—for 235 bio-based chemicals. Using GEMs, the researchers calculated both the maximum theoretical yields and the maximum achievable yields under industrial conditions for each chemical, thereby establishing criteria to identify the most suitable strains for each target compound.
< Figure 1. Outline of the strategy for improving microbial cell factories using a genome-scale metabolic model (GEM) >
The team specifically proposed strategies such as introducing heterologous enzyme reactions derived from other organisms and exchanging cofactors used by microbes to expand metabolic pathways. These strategies were shown to increase yields beyond the innate metabolic capacities of the microorganisms, resulting in higher production of industrially important chemicals such as mevalonic acid, propanol, fatty acids, and isoprenoids.
Moreover, by applying a computational approach to analyze metabolic fluxes in silico, the researchers suggested strategies for improving microbial strains to maximize the production of various chemicals. They quantitatively identified the relationships between specific enzyme reactions and target chemical production, as well as the relationships between enzymes and metabolites, determining which enzyme reactions should be up- or down-regulated. Through this, the team presented strategies not only to achieve high theoretical yields but also to maximize actual production capacities.
< Figure 2. Comparison of production routes and maximum yields of useful chemicals using representative industrial microorganisms >
Dr. Gi Bae Kim, the first author of this paper from the KAIST BioProcess Engineering Research Center, explained, “By introducing metabolic pathways derived from other organisms and exchanging cofactors, it is possible to design new microbial cell factories that surpass existing limitations. The strategies presented in this study will play a pivotal role in making microbial-based production processes more economical and efficient.” In addition, Distinguished Professor Sang Yup Lee noted, “This research serves as a key resource in the field of systems metabolic engineering, reducing difficulties in strain selection and pathway design, and enabling more efficient development of microbial cell factories. We expect it to greatly contribute to the future development of technologies for producing various eco-friendly chemicals, such as biofuels, bioplastics, and functional food materials.”
This research was conducted with the support from the Development of platform technologies of microbial cell factories for the next-generation biorefineries project and Development of advanced synthetic biology source technologies for leading the biomanufacturing industry project (Project Leader: Distinguished Professor Sang Yup Lee, KAIST) from National Research Foundation supported by the Korean Ministry of Science and ICT.
2025.03.27 View 3402
KAIST provides a comprehensive resource on microbial cell factories for sustainable chemical production
In silico analysis of five industrial microorganisms identifies optimal strains and metabolic engineering strategies for producing 235 valuable chemicals
Climate change and the depletion of fossil fuels have raised the global need for sustainable chemical production. In response to these environmental challenges, microbial cell factories are gaining attention as eco-friendly platforms for producing chemicals using renewable resources, while metabolic engineering technologies to enhance these cell factories are becoming crucial tools for maximizing production efficiency. However, difficulties in selecting suitable microbial strains and optimizing complex metabolic pathways continue to pose significant obstacles to practical industrial applications.
KAIST (President Kwang-Hyung Lee) announced on 27th of March that Distinguished Professor Sang Yup Lee’s research team in the Department of Chemical and Biomolecular Engineering comprehensively evaluated the production capabilities of various industrial microbial cell factories using in silico simulations and, based on these findings, identified the most suitable microbial strains for producing specific chemicals as well as optimal metabolic engineering strategies.
Previously, researchers attempted to determine the best strains and efficient metabolic engineering strategies among numerous microbial candidates through extensive biological experiments and meticulous verification processes. However, this approach required substantial time and costs. Recently, the introduction of genome-scale metabolic models (GEMs), which reconstruct the metabolic networks within an organism based on its entire genome information, has enabled systematic analysis of metabolic fluxes via computer simulations. This development offers a new way to overcome limitations of conventional experimental approaches, revolutionizing both strain selection and metabolic pathway design.
Accordingly, Professor Lee’s team at the Department of Chemical and Biomolecular Engineering, KAIST, evaluated the production capabilities of five representative industrial microorganisms—Escherichia coli, Saccharomyces cerevisiae, Bacillus subtilis, Corynebacterium glutamicum, and Pseudomonas putida—for 235 bio-based chemicals. Using GEMs, the researchers calculated both the maximum theoretical yields and the maximum achievable yields under industrial conditions for each chemical, thereby establishing criteria to identify the most suitable strains for each target compound.
< Figure 1. Outline of the strategy for improving microbial cell factories using a genome-scale metabolic model (GEM) >
The team specifically proposed strategies such as introducing heterologous enzyme reactions derived from other organisms and exchanging cofactors used by microbes to expand metabolic pathways. These strategies were shown to increase yields beyond the innate metabolic capacities of the microorganisms, resulting in higher production of industrially important chemicals such as mevalonic acid, propanol, fatty acids, and isoprenoids.
Moreover, by applying a computational approach to analyze metabolic fluxes in silico, the researchers suggested strategies for improving microbial strains to maximize the production of various chemicals. They quantitatively identified the relationships between specific enzyme reactions and target chemical production, as well as the relationships between enzymes and metabolites, determining which enzyme reactions should be up- or down-regulated. Through this, the team presented strategies not only to achieve high theoretical yields but also to maximize actual production capacities.
< Figure 2. Comparison of production routes and maximum yields of useful chemicals using representative industrial microorganisms >
Dr. Gi Bae Kim, the first author of this paper from the KAIST BioProcess Engineering Research Center, explained, “By introducing metabolic pathways derived from other organisms and exchanging cofactors, it is possible to design new microbial cell factories that surpass existing limitations. The strategies presented in this study will play a pivotal role in making microbial-based production processes more economical and efficient.” In addition, Distinguished Professor Sang Yup Lee noted, “This research serves as a key resource in the field of systems metabolic engineering, reducing difficulties in strain selection and pathway design, and enabling more efficient development of microbial cell factories. We expect it to greatly contribute to the future development of technologies for producing various eco-friendly chemicals, such as biofuels, bioplastics, and functional food materials.”
This research was conducted with the support from the Development of platform technologies of microbial cell factories for the next-generation biorefineries project and Development of advanced synthetic biology source technologies for leading the biomanufacturing industry project (Project Leader: Distinguished Professor Sang Yup Lee, KAIST) from National Research Foundation supported by the Korean Ministry of Science and ICT.
2025.03.27 View 3402 -
 KAIST Innovates Mid-Infrared Photodetectors for Exoplanet Detection, Expanding Applications to Environmental and Medical Fields
NASA’s James Webb Space Telescope (JWST) utilizes mid-infrared spectroscopy to precisely analyze molecular components such as water vapor and sulfur dioxide in exoplanet atmospheres. The key to this analysis, where each molecule exhibits a unique spectral "fingerprint," lies in highly sensitive photodetector technology capable of measuring extremely weak light intensities. Recently, KAIST researchers have developed an innovative photodetector capable of detecting a broad range of mid-infrared spectra, garnering significant attention.
< Photo 1. (from the left) Ph.D. candidate Inki Kim (co-author), Professor SangHyeon Kim (corresponding author), Dr. Joonsup Shim (first author), and Dr. Jinha Lim (co-author) of KAIST School of Electrical Engineering. >
KAIST (represented by President Kwang-Hyung Lee) announced on the 27th of March that a research team led by Professor SangHyeon Kim from the School of Electrical Engineering has developed a mid-infrared photodetector that operates stably at room temperature, marking a major turning point for the commercialization of ultra-compact optical sensors.
The newly developed photodetector utilizes conventional silicon-based CMOS processes, enabling low-cost mass production while maintaining stable operation at room temperature. Notably, the research team successfully demonstrated the real-time detection of carbon dioxide (CO₂) gas using ultra-compact and ultra-thin optical sensors equipped with this photodetector, proving its potential for environmental monitoring and hazardous gas analysis.
Existing mid-infrared photodetectors generally require cooling systems due to high thermal noise at room temperature. These cooling systems increase the size and cost of equipment, making miniaturization and integration into portable devices challenging. Furthermore, conventional mid-infrared photodetectors are incompatible with silicon-based CMOS processes, limiting large-scale production and commercialization.
To address these limitations, the research team developed a waveguide-integrated photodetector using germanium (Ge), a Group IV element like silicon. This approach enables broad-spectrum mid-infrared detection while ensuring stable operation at room temperature.
< Figure 1. Schematic diagram of a room-temperature mid-infrared waveguide-integrated photodetector based on the Ge-on-insulator optical platform proposed in this study (top). Optical microscope image of the integrated photodetector connected with the sensing unit (bottom). >
A waveguide is a structure designed to efficiently guide light along a specific path with minimal loss. To implement various optical functions on a chip (on-chip), the development of waveguide-integrated photodetectors and waveguide-based optical components is essential.
Unlike conventional photodetectors that primarily rely on bandgap absorption principles, this new technology leverages the bolometric effect*, allowing it to detect the entire mid-infrared spectral range. As a result, it can be widely applied to the real-time sensing of various molecular species.
*Bolometric effect: A principle in which light absorption leads to an increase in temperature, causing electrical signals to change accordingly.
The waveguide-integrated mid-infrared photodetector developed by the research team is considered a groundbreaking innovation that overcomes the limitations of existing mid-infrared sensor technologies, including the need for cooling, difficulties in mass production, and high costs.
< Figure 2. Room temperature photoresponse characteristics of the mid-infrared waveguide photodetector proposed in this study (left) and real-time carbon dioxide (CO2) gas sensing results using the photodetector (right). >
This breakthrough technology is expected to be applicable across diverse fields, including environmental monitoring, medical diagnostics, industrial process management, national defense and security, and smart devices. It also paves the way for next-generation mid-infrared sensor advancements.
Professor SangHyeon Kim from KAIST stated, "This research represents a novel approach that overcomes the limitations of existing mid-infrared photodetector technologies and has great potential for practical applications in various fields." He further emphasized, "Since this sensor technology is compatible with CMOS processes, it enables low-cost mass production, making it highly suitable for next-generation environmental monitoring systems and smart manufacturing sites."
< Figure 3. Performance comparison image of a room-temperature mid-infrared waveguide photodetector fabricated with the technology proposed in this study. It achieves the world’s highest performance compared to existing technologies utilizing the Bolometric effect, and is the only solution compatible with CMOS processes. The technology proposed by our research team is characterized by its ability to respond to a wide spectrum of the mid-infrared band without limitations. >
The study, with Dr. Joonsup Shim (currently a postdoctoral researcher at Harvard University) as the first author, was published on March 19, 2025 in the internationally renowned journal Light: Science & Applications (JCR 2.9%, IF=20.6).
(Paper title: “Room-temperature waveguide-integrated photodetector using bolometric effect for mid-infrared spectroscopy applications,” https://doi.org/10.1038/s41377-025-01803-3)
2025.03.27 View 2381
KAIST Innovates Mid-Infrared Photodetectors for Exoplanet Detection, Expanding Applications to Environmental and Medical Fields
NASA’s James Webb Space Telescope (JWST) utilizes mid-infrared spectroscopy to precisely analyze molecular components such as water vapor and sulfur dioxide in exoplanet atmospheres. The key to this analysis, where each molecule exhibits a unique spectral "fingerprint," lies in highly sensitive photodetector technology capable of measuring extremely weak light intensities. Recently, KAIST researchers have developed an innovative photodetector capable of detecting a broad range of mid-infrared spectra, garnering significant attention.
< Photo 1. (from the left) Ph.D. candidate Inki Kim (co-author), Professor SangHyeon Kim (corresponding author), Dr. Joonsup Shim (first author), and Dr. Jinha Lim (co-author) of KAIST School of Electrical Engineering. >
KAIST (represented by President Kwang-Hyung Lee) announced on the 27th of March that a research team led by Professor SangHyeon Kim from the School of Electrical Engineering has developed a mid-infrared photodetector that operates stably at room temperature, marking a major turning point for the commercialization of ultra-compact optical sensors.
The newly developed photodetector utilizes conventional silicon-based CMOS processes, enabling low-cost mass production while maintaining stable operation at room temperature. Notably, the research team successfully demonstrated the real-time detection of carbon dioxide (CO₂) gas using ultra-compact and ultra-thin optical sensors equipped with this photodetector, proving its potential for environmental monitoring and hazardous gas analysis.
Existing mid-infrared photodetectors generally require cooling systems due to high thermal noise at room temperature. These cooling systems increase the size and cost of equipment, making miniaturization and integration into portable devices challenging. Furthermore, conventional mid-infrared photodetectors are incompatible with silicon-based CMOS processes, limiting large-scale production and commercialization.
To address these limitations, the research team developed a waveguide-integrated photodetector using germanium (Ge), a Group IV element like silicon. This approach enables broad-spectrum mid-infrared detection while ensuring stable operation at room temperature.
< Figure 1. Schematic diagram of a room-temperature mid-infrared waveguide-integrated photodetector based on the Ge-on-insulator optical platform proposed in this study (top). Optical microscope image of the integrated photodetector connected with the sensing unit (bottom). >
A waveguide is a structure designed to efficiently guide light along a specific path with minimal loss. To implement various optical functions on a chip (on-chip), the development of waveguide-integrated photodetectors and waveguide-based optical components is essential.
Unlike conventional photodetectors that primarily rely on bandgap absorption principles, this new technology leverages the bolometric effect*, allowing it to detect the entire mid-infrared spectral range. As a result, it can be widely applied to the real-time sensing of various molecular species.
*Bolometric effect: A principle in which light absorption leads to an increase in temperature, causing electrical signals to change accordingly.
The waveguide-integrated mid-infrared photodetector developed by the research team is considered a groundbreaking innovation that overcomes the limitations of existing mid-infrared sensor technologies, including the need for cooling, difficulties in mass production, and high costs.
< Figure 2. Room temperature photoresponse characteristics of the mid-infrared waveguide photodetector proposed in this study (left) and real-time carbon dioxide (CO2) gas sensing results using the photodetector (right). >
This breakthrough technology is expected to be applicable across diverse fields, including environmental monitoring, medical diagnostics, industrial process management, national defense and security, and smart devices. It also paves the way for next-generation mid-infrared sensor advancements.
Professor SangHyeon Kim from KAIST stated, "This research represents a novel approach that overcomes the limitations of existing mid-infrared photodetector technologies and has great potential for practical applications in various fields." He further emphasized, "Since this sensor technology is compatible with CMOS processes, it enables low-cost mass production, making it highly suitable for next-generation environmental monitoring systems and smart manufacturing sites."
< Figure 3. Performance comparison image of a room-temperature mid-infrared waveguide photodetector fabricated with the technology proposed in this study. It achieves the world’s highest performance compared to existing technologies utilizing the Bolometric effect, and is the only solution compatible with CMOS processes. The technology proposed by our research team is characterized by its ability to respond to a wide spectrum of the mid-infrared band without limitations. >
The study, with Dr. Joonsup Shim (currently a postdoctoral researcher at Harvard University) as the first author, was published on March 19, 2025 in the internationally renowned journal Light: Science & Applications (JCR 2.9%, IF=20.6).
(Paper title: “Room-temperature waveguide-integrated photodetector using bolometric effect for mid-infrared spectroscopy applications,” https://doi.org/10.1038/s41377-025-01803-3)
2025.03.27 View 2381 -
 KAIST Captures Protein Reaction in Just Six Milliseconds
Understanding biomolecular processes - such as protein-protein interactions and enzyme-substrate reactions that occur on the microseconds to millisecond time scale is essential for comprehending life processes and advancing drug development. KAIST researchers have developed a method for freezing and analyzing biochemical reaction dynamics within a span of just a few milliseconds, marking a significant step forward in better understanding complex biological reactions.
< Photo. (From left) Professor Jin Young Kang and Haerang Hwang of the Integrated Master's and Doctoral Program of the Department of Chemistry, along with Professor Wonhee Lee of the Department of Physics >
KAIST (represented by President Kwang Hyung Lee) announced on the 24th of March that a joint research team led by Professor Jin Young Kang from the Department of Chemistry and Professor Wonhee Lee from the Department of Physics has developed a parylene-based thin-film microfluidic mixing-and-spraying device for ultra-fast biochemical reaction studies.
*Parylene: A key material for microfluidic devices used to observe protein dynamics at ultra-high speeds. It can be fabricated into a few micrometer-thick films, which can be used in making a spray nozzle for microfluidic devices.
This research overcomes the limitations of the existing time-resolved cryo-electron microscopy (TRCEM) method by reducing sample consumption to one-third of the conventional amount while improving the minimum time resolution—down to just six milliseconds (6 ms).
TRCEM is a technique that rapidly freezes protein complexes during intermediate reaction stages under cryogenic conditions, which allows researchers to analyze their structures. This approach has gained significant attention recently for its ability to capture transient biochemical events.
< Figure 1. Time-resolved cryo-EM (TRCEM) technique using microfluidic channels. In order to capture the intermediate structure of biomolecules during a biochemical reaction over time, biomolecules and reaction substrates are mixed in a microfluidic channel, and then sprayed on a grid after a certain reaction time and frozen in liquid ethane to prepare a cryo-EM sample. This can then be analyzed by cryo-EM to observe the structural changes of proteins over time. >
Transient intermediate structures of protein complexes could not be captured by traditional cryo-electron microscopy due to their extremely short lifespans. Although several TRCEM techniques have been developed to address this issue, previous methods were hindered by large sample consumption and limited time resolution. To overcome these challenges, the KAIST team developed a new mixing-and-spraying device using ultra-thin parylene films. The integrated design of the device further enhanced the precision and reproducibility of experiments.
< Figure 2. TRCEM grid fabrication setup using a parylene-based thin-film microfluidic device and actual appearance of the device. You can see that a thin-film parylene channel is inserted into the injection nozzle. The integration of the reaction channel and the injection nozzle allowed the residence time in the device to be reduced to at least 0.5 ms. >
“This research makes TRCEM more practical and paves the way for diverse applications of the parylene thin-film device in structural biology, drug development, enzyme reaction studies, and biosensor research.” Professor Jin Young Kang explained, emphasizing the significance of the study.
Professor Wonhee Lee added, “The team aims to continue this research, focusing on improvement of the technique to achieve higher time resolution with minimal sample consumption.”
< Figure 3. Comparison of the spraying patterns of the parylene mixing-jet device and the conventional mixing-jet device and the filament length in the resulting RecA-ssDNA filament formation reaction. It was shown that the thin film spray nozzle structure affects the uniformity and accuracy of the final reaction time. >
The research findings, with Haerang Hwang (a graduate student in the integrated master's and Ph.D. program in the Department of Chemistry) as the first author, were published online on January 28, 2025, in the international journal Advanced Functional Materials. (Paper Title: “Integrated Parylene-Based Thin-Film Microfluidic Device for Time-Resolved Cryo-Electron Microscopy”, DOI: doi.org/10.1002/adfm.202418224)
This research was supported by the National Research Foundation of Korea (NRF), the Samsung Future Technology Development Program, and the CELINE consortium.
2025.03.24 View 3228
KAIST Captures Protein Reaction in Just Six Milliseconds
Understanding biomolecular processes - such as protein-protein interactions and enzyme-substrate reactions that occur on the microseconds to millisecond time scale is essential for comprehending life processes and advancing drug development. KAIST researchers have developed a method for freezing and analyzing biochemical reaction dynamics within a span of just a few milliseconds, marking a significant step forward in better understanding complex biological reactions.
< Photo. (From left) Professor Jin Young Kang and Haerang Hwang of the Integrated Master's and Doctoral Program of the Department of Chemistry, along with Professor Wonhee Lee of the Department of Physics >
KAIST (represented by President Kwang Hyung Lee) announced on the 24th of March that a joint research team led by Professor Jin Young Kang from the Department of Chemistry and Professor Wonhee Lee from the Department of Physics has developed a parylene-based thin-film microfluidic mixing-and-spraying device for ultra-fast biochemical reaction studies.
*Parylene: A key material for microfluidic devices used to observe protein dynamics at ultra-high speeds. It can be fabricated into a few micrometer-thick films, which can be used in making a spray nozzle for microfluidic devices.
This research overcomes the limitations of the existing time-resolved cryo-electron microscopy (TRCEM) method by reducing sample consumption to one-third of the conventional amount while improving the minimum time resolution—down to just six milliseconds (6 ms).
TRCEM is a technique that rapidly freezes protein complexes during intermediate reaction stages under cryogenic conditions, which allows researchers to analyze their structures. This approach has gained significant attention recently for its ability to capture transient biochemical events.
< Figure 1. Time-resolved cryo-EM (TRCEM) technique using microfluidic channels. In order to capture the intermediate structure of biomolecules during a biochemical reaction over time, biomolecules and reaction substrates are mixed in a microfluidic channel, and then sprayed on a grid after a certain reaction time and frozen in liquid ethane to prepare a cryo-EM sample. This can then be analyzed by cryo-EM to observe the structural changes of proteins over time. >
Transient intermediate structures of protein complexes could not be captured by traditional cryo-electron microscopy due to their extremely short lifespans. Although several TRCEM techniques have been developed to address this issue, previous methods were hindered by large sample consumption and limited time resolution. To overcome these challenges, the KAIST team developed a new mixing-and-spraying device using ultra-thin parylene films. The integrated design of the device further enhanced the precision and reproducibility of experiments.
< Figure 2. TRCEM grid fabrication setup using a parylene-based thin-film microfluidic device and actual appearance of the device. You can see that a thin-film parylene channel is inserted into the injection nozzle. The integration of the reaction channel and the injection nozzle allowed the residence time in the device to be reduced to at least 0.5 ms. >
“This research makes TRCEM more practical and paves the way for diverse applications of the parylene thin-film device in structural biology, drug development, enzyme reaction studies, and biosensor research.” Professor Jin Young Kang explained, emphasizing the significance of the study.
Professor Wonhee Lee added, “The team aims to continue this research, focusing on improvement of the technique to achieve higher time resolution with minimal sample consumption.”
< Figure 3. Comparison of the spraying patterns of the parylene mixing-jet device and the conventional mixing-jet device and the filament length in the resulting RecA-ssDNA filament formation reaction. It was shown that the thin film spray nozzle structure affects the uniformity and accuracy of the final reaction time. >
The research findings, with Haerang Hwang (a graduate student in the integrated master's and Ph.D. program in the Department of Chemistry) as the first author, were published online on January 28, 2025, in the international journal Advanced Functional Materials. (Paper Title: “Integrated Parylene-Based Thin-Film Microfluidic Device for Time-Resolved Cryo-Electron Microscopy”, DOI: doi.org/10.1002/adfm.202418224)
This research was supported by the National Research Foundation of Korea (NRF), the Samsung Future Technology Development Program, and the CELINE consortium.
2025.03.24 View 3228