the+National+Academy+of+Sciences
-
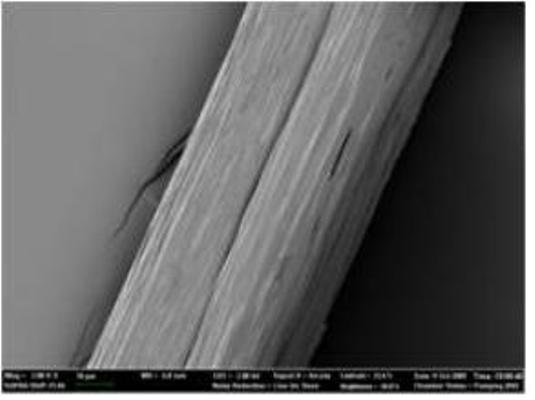 Native-like Spider Silk Produced in Metabolically Engineered Bacterium
Microscopic picture of 285 kilodalton recombinant spider silk fiber
Researchers have long envied spiders’ ability to manufacture silk that is light-weighted while as strong and tough as steel or Kevlar. Indeed, finer than human hair, five times stronger by weight than steel, and three times tougher than the top quality man-made fiber Kevlar, spider dragline silk is an ideal material for numerous applications. Suggested industrial applications have ranged from parachute cords and protective clothing to composite materials in aircrafts. Also, many biomedical applications are envisioned due to its biocompatibility and biodegradability.
Unfortunately, natural dragline silk cannot be conveniently obtained by farming spiders because they are highly territorial and aggressive. To develop a more sustainable process, can scientists mass-produce artificial silk while maintaining the amazing properties of native silk? That is something Sang Yup Lee at the Korea Advanced Institute of Science and Technology (KAIST) in Daejeon, the Republic of Korea, and his collaborators, Professor Young Hwan Park at Seoul National University and Professor David Kaplan at Tufts University, wanted to figure out. Their method is very similar to what spiders essentially do: first, expression of recombinant silk proteins; second, making the soluble silk proteins into water-insoluble fibers through spinning.
For the successful expression of high molecular weight spider silk protein, Professor Lee and his colleagues pieced together the silk gene from chemically synthesized oligonucleotides, and then inserted it into the expression host (in this case, an industrially safe bacterium Escherichia coli which is normally found in our gut). Initially, the bacterium refused to the challenging task of producing high molecular weight spider silk protein due to the unique characteristics of the protein, such as extremely large size, repetitive nature of the protein structure, and biased abundance of a particular amino acid glycine. “To make E. coli synthesize this ultra high molecular weight (as big as 285 kilodalton) spider silk protein having highly repetitive amino acid sequence, we helped E. coli overcome the difficulties by systems metabolic engineering,” says Sang Yup Lee, Distinguished Professor of KAIST, who led this project. His team boosted the pool of glycyl-tRNA, the major building block of spider silk protein synthesis. “We could obtain appreciable expression of the 285 kilodalton spider silk protein, which is the largest recombinant silk protein ever produced in E. coli. That was really incredible.” says Dr. Xia.
But this was only step one. The KAIST team performed high-cell-density cultures for mass production of the recombinant spider silk protein. Then, the team developed a simple, easy to scale-up purification process for the recombinant spider silk protein. The purified spider silk protein could be spun into beautiful silk fiber. To study the mechanical properties of the artificial spider silk, the researchers determined tenacity, elongation, and Young’s modulus, the three critical mechanical parameters that represent a fiber’s strength, extensibility, and stiffness. Importantly, the artificial fiber displayed the tenacity, elongation, and Young’s modulus of 508 MPa, 15%, and 21 GPa, respectively, which are comparable to those of the native spider silk.
“We have offered an overall platform for mass production of native-like spider dragline silk. This platform would enable us to have broader industrial and biomedical applications for spider silk. Moreover, many other silk-like biomaterials such as elastin, collagen, byssus, resilin, and other repetitive proteins have similar features to spider silk protein. Thus, our platform should also be useful for their efficient bio-based production and applications,” concludes Professor Lee.
This work is published on July 26 in the Proceedings of the National Academy of Sciences (PNAS) online.
2010.07.28 View 21981
Native-like Spider Silk Produced in Metabolically Engineered Bacterium
Microscopic picture of 285 kilodalton recombinant spider silk fiber
Researchers have long envied spiders’ ability to manufacture silk that is light-weighted while as strong and tough as steel or Kevlar. Indeed, finer than human hair, five times stronger by weight than steel, and three times tougher than the top quality man-made fiber Kevlar, spider dragline silk is an ideal material for numerous applications. Suggested industrial applications have ranged from parachute cords and protective clothing to composite materials in aircrafts. Also, many biomedical applications are envisioned due to its biocompatibility and biodegradability.
Unfortunately, natural dragline silk cannot be conveniently obtained by farming spiders because they are highly territorial and aggressive. To develop a more sustainable process, can scientists mass-produce artificial silk while maintaining the amazing properties of native silk? That is something Sang Yup Lee at the Korea Advanced Institute of Science and Technology (KAIST) in Daejeon, the Republic of Korea, and his collaborators, Professor Young Hwan Park at Seoul National University and Professor David Kaplan at Tufts University, wanted to figure out. Their method is very similar to what spiders essentially do: first, expression of recombinant silk proteins; second, making the soluble silk proteins into water-insoluble fibers through spinning.
For the successful expression of high molecular weight spider silk protein, Professor Lee and his colleagues pieced together the silk gene from chemically synthesized oligonucleotides, and then inserted it into the expression host (in this case, an industrially safe bacterium Escherichia coli which is normally found in our gut). Initially, the bacterium refused to the challenging task of producing high molecular weight spider silk protein due to the unique characteristics of the protein, such as extremely large size, repetitive nature of the protein structure, and biased abundance of a particular amino acid glycine. “To make E. coli synthesize this ultra high molecular weight (as big as 285 kilodalton) spider silk protein having highly repetitive amino acid sequence, we helped E. coli overcome the difficulties by systems metabolic engineering,” says Sang Yup Lee, Distinguished Professor of KAIST, who led this project. His team boosted the pool of glycyl-tRNA, the major building block of spider silk protein synthesis. “We could obtain appreciable expression of the 285 kilodalton spider silk protein, which is the largest recombinant silk protein ever produced in E. coli. That was really incredible.” says Dr. Xia.
But this was only step one. The KAIST team performed high-cell-density cultures for mass production of the recombinant spider silk protein. Then, the team developed a simple, easy to scale-up purification process for the recombinant spider silk protein. The purified spider silk protein could be spun into beautiful silk fiber. To study the mechanical properties of the artificial spider silk, the researchers determined tenacity, elongation, and Young’s modulus, the three critical mechanical parameters that represent a fiber’s strength, extensibility, and stiffness. Importantly, the artificial fiber displayed the tenacity, elongation, and Young’s modulus of 508 MPa, 15%, and 21 GPa, respectively, which are comparable to those of the native spider silk.
“We have offered an overall platform for mass production of native-like spider dragline silk. This platform would enable us to have broader industrial and biomedical applications for spider silk. Moreover, many other silk-like biomaterials such as elastin, collagen, byssus, resilin, and other repetitive proteins have similar features to spider silk protein. Thus, our platform should also be useful for their efficient bio-based production and applications,” concludes Professor Lee.
This work is published on July 26 in the Proceedings of the National Academy of Sciences (PNAS) online.
2010.07.28 View 21981 -
 Scaling Laws between Population and Facility Densities Found
A research team led by Prof. Ha-Woong Jeong of the Department of Physics, KAIST, has found a positive correlation between facilities and population densities, university authorities said on Tuesday (Sept. 2). The research was conducted in the cooperation with a research team of Prof. Beom-Jun Kim at Sungkyunkwan University.
The researchers investigated the ideal relation between the population and the facilities within the framework of an economic mechanism governing microdynamics.
In previous studies based on the global optimization of facility positions in minimizing the overall travel distance between people and facilities, the relation between population and facilities should follow a simple law. The new empirical analysis, however, determined that the law is not a fixed value but spreads in a broad range depending on facility types.
To explain this discrepancy, the researchers proposed a model based on economic mechanism that mimics the competitive balance between the profit of the facilities and the social opportunity cost for population.
The results were published in the Proceedings of the National Academy of Sciences of the United States on Aug. 25.
2009.09.04 View 16362
Scaling Laws between Population and Facility Densities Found
A research team led by Prof. Ha-Woong Jeong of the Department of Physics, KAIST, has found a positive correlation between facilities and population densities, university authorities said on Tuesday (Sept. 2). The research was conducted in the cooperation with a research team of Prof. Beom-Jun Kim at Sungkyunkwan University.
The researchers investigated the ideal relation between the population and the facilities within the framework of an economic mechanism governing microdynamics.
In previous studies based on the global optimization of facility positions in minimizing the overall travel distance between people and facilities, the relation between population and facilities should follow a simple law. The new empirical analysis, however, determined that the law is not a fixed value but spreads in a broad range depending on facility types.
To explain this discrepancy, the researchers proposed a model based on economic mechanism that mimics the competitive balance between the profit of the facilities and the social opportunity cost for population.
The results were published in the Proceedings of the National Academy of Sciences of the United States on Aug. 25.
2009.09.04 View 16362 -
 Maximum Yield Amino Acid-Producing Microorganism Developed with use of System Biotechnology
Maximum Yield Amino Acid-Producing Microorganism Developed with use of System Biotechnology
A team led by Sang-Yup Lee, a distinguished professor of Chemical and Biomolecular Engineering and chair professor of LG Chemical, has succeeded in developing maximum yield L-valine-producing microorganism by using System Biotechnology methods. The research results will be published at the April fourth week (April 23 - 27) edition of the Proceedings of the National Academy of Sciences (PNAS) of the USA.
Prof. Lee’s team has developed maximum yield amino acid-producing microorganism (target substance of L-valine, an essential amino-acid) by using microorganism E cell system and simulation methods.
His team produced initial producing microorganism by selectively operating necessary parts in colon bacillus genome and excavated preliminary target gene which is to newly be operated through transcriptome analysis using DNA chips. Then they performed a great amount of gene deletion experiment on computer by using MBEL979, E-cells of colon bacillus, and excavated secondary engineering targets. And they finally succeeded in developing maximum yield valine-producing microorganism that can extract 37.8 grams of valine from 100 grams of glucose by applying experiment results to the actual development of microorganism so as to achieve the optimization of metabolic flux in cells,
Prof. Lee said, “Since successfully used for the development of microorganism on a systematic system level, system biotechnology methods are expected to significantly contribute to the development of all biotechnology-relevant industries. At the beginning, we had huge obstacles in fusing IT and BT, but my team mates cleverly overcame such obstacles, hence I’m very proud of them.” The producing microorganism and its developing methods are pending international applications (PCT).
2007.04.26 View 17877
Maximum Yield Amino Acid-Producing Microorganism Developed with use of System Biotechnology
Maximum Yield Amino Acid-Producing Microorganism Developed with use of System Biotechnology
A team led by Sang-Yup Lee, a distinguished professor of Chemical and Biomolecular Engineering and chair professor of LG Chemical, has succeeded in developing maximum yield L-valine-producing microorganism by using System Biotechnology methods. The research results will be published at the April fourth week (April 23 - 27) edition of the Proceedings of the National Academy of Sciences (PNAS) of the USA.
Prof. Lee’s team has developed maximum yield amino acid-producing microorganism (target substance of L-valine, an essential amino-acid) by using microorganism E cell system and simulation methods.
His team produced initial producing microorganism by selectively operating necessary parts in colon bacillus genome and excavated preliminary target gene which is to newly be operated through transcriptome analysis using DNA chips. Then they performed a great amount of gene deletion experiment on computer by using MBEL979, E-cells of colon bacillus, and excavated secondary engineering targets. And they finally succeeded in developing maximum yield valine-producing microorganism that can extract 37.8 grams of valine from 100 grams of glucose by applying experiment results to the actual development of microorganism so as to achieve the optimization of metabolic flux in cells,
Prof. Lee said, “Since successfully used for the development of microorganism on a systematic system level, system biotechnology methods are expected to significantly contribute to the development of all biotechnology-relevant industries. At the beginning, we had huge obstacles in fusing IT and BT, but my team mates cleverly overcame such obstacles, hence I’m very proud of them.” The producing microorganism and its developing methods are pending international applications (PCT).
2007.04.26 View 17877