National+Research+Foundation
-
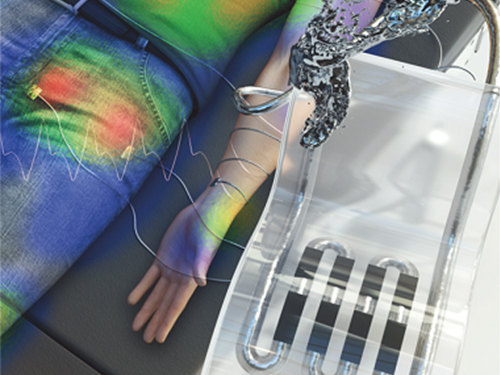 New Liquid Metal Wearable Pressure Sensor Created for Health Monitoring Applications
Soft pressure sensors have received significant research attention in a variety of fields, including soft robotics, electronic skin, and wearable electronics. Wearable soft pressure sensors have great potential for the real-time health monitoring and for the early diagnosis of diseases.
A KAIST research team led by Professor Inkyu Park from the Department of Mechanical Engineering developed a highly sensitive wearable pressure sensor for health monitoring applications. This work was reported in Advanced Healthcare Materials on November 21 as a front cover article.
This technology is capable of sensitive, precise, and continuous measurement of physiological and physical signals and shows great potential for health monitoring applications and the early diagnosis of diseases.
A soft pressure sensor is required to have high compliance, high sensitivity, low cost, long-term performance stability, and environmental stability in order to be employed for continuous health monitoring. Conventional solid-state soft pressure sensors using functional materials including carbon nanotubes and graphene have showed great sensing performance. However, these sensors suffer from limited stretchability, signal drifting, and long-term instability due to the distance between the stretchable substrate and the functional materials.
To overcome these issues, liquid-state electronics using liquid metal have been introduced for various wearable applications. Of these materials, Galinstan, a eutectic metal alloy of gallium, indium, and tin, has great mechanical and electrical properties that can be employed in wearable applications. But today’s liquid metal-based pressure sensors have low-pressure sensitivity, limiting their applicability for health monitoring devices.
The research team developed a 3D-printed rigid microbump array-integrated, liquid metal-based soft pressure sensor. With the help of 3D printing, the integration of a rigid microbump array and the master mold for a liquid metal microchannel could be achieved simultaneously, reducing the complexity of the manufacturing process. Through the integration of the rigid microbump and the microchannel, the new pressure sensor has an extremely low detection limit and enhanced pressure sensitivity compared to previously reported liquid metal-based pressure sensors. The proposed sensor also has a negligible signal drift over 10,000 cycles of pressure, bending, and stretching and exhibited excellent stability when subjected to various environmental conditions.
These performance outcomes make it an excellent sensor for various health monitoring devices. First, the research team demonstrated a wearable wristband device that can continuously monitor one’s pulse during exercise and be employed in a noninvasive cuffless BP monitoring system based on PTT calculations. Then, they introduced a wireless wearable heel pressure monitoring system that integrates three 3D-BLiPS with a wireless communication module.
Professor Park said, “It was possible to measure health indicators including pulse and blood pressure continuously as well as pressure of body parts using our proposed soft pressure sensor. We expect it to be used in health care applications, such as the prevention and the monitoring of the pressure-driven diseases such as pressure ulcers in the near future. There will be more opportunities for future research including a whole-body pressure monitoring system related to other physical parameters.”
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Ministry of Science and ICT.
< Figure 1. The front cover image of Advanced Healthcare Materials, Volume 8, Issue 22. >
< Figure 2. Highly sensitive liquid metal-based soft pressure sensor integrated with 3D-printed microbump array. >
< Figure 3. High pressure sensitivity and reliable sensing performances of the proposed sensor and wireless heel pressure monitoring application. >
-ProfileProfessor Inkyu ParkMicro/Nano Transducers Laboratoryhttp://mintlab1.kaist.ac.kr/
Department of Mechanical EngineeringKAIST
2019.12.20 View 17571
New Liquid Metal Wearable Pressure Sensor Created for Health Monitoring Applications
Soft pressure sensors have received significant research attention in a variety of fields, including soft robotics, electronic skin, and wearable electronics. Wearable soft pressure sensors have great potential for the real-time health monitoring and for the early diagnosis of diseases.
A KAIST research team led by Professor Inkyu Park from the Department of Mechanical Engineering developed a highly sensitive wearable pressure sensor for health monitoring applications. This work was reported in Advanced Healthcare Materials on November 21 as a front cover article.
This technology is capable of sensitive, precise, and continuous measurement of physiological and physical signals and shows great potential for health monitoring applications and the early diagnosis of diseases.
A soft pressure sensor is required to have high compliance, high sensitivity, low cost, long-term performance stability, and environmental stability in order to be employed for continuous health monitoring. Conventional solid-state soft pressure sensors using functional materials including carbon nanotubes and graphene have showed great sensing performance. However, these sensors suffer from limited stretchability, signal drifting, and long-term instability due to the distance between the stretchable substrate and the functional materials.
To overcome these issues, liquid-state electronics using liquid metal have been introduced for various wearable applications. Of these materials, Galinstan, a eutectic metal alloy of gallium, indium, and tin, has great mechanical and electrical properties that can be employed in wearable applications. But today’s liquid metal-based pressure sensors have low-pressure sensitivity, limiting their applicability for health monitoring devices.
The research team developed a 3D-printed rigid microbump array-integrated, liquid metal-based soft pressure sensor. With the help of 3D printing, the integration of a rigid microbump array and the master mold for a liquid metal microchannel could be achieved simultaneously, reducing the complexity of the manufacturing process. Through the integration of the rigid microbump and the microchannel, the new pressure sensor has an extremely low detection limit and enhanced pressure sensitivity compared to previously reported liquid metal-based pressure sensors. The proposed sensor also has a negligible signal drift over 10,000 cycles of pressure, bending, and stretching and exhibited excellent stability when subjected to various environmental conditions.
These performance outcomes make it an excellent sensor for various health monitoring devices. First, the research team demonstrated a wearable wristband device that can continuously monitor one’s pulse during exercise and be employed in a noninvasive cuffless BP monitoring system based on PTT calculations. Then, they introduced a wireless wearable heel pressure monitoring system that integrates three 3D-BLiPS with a wireless communication module.
Professor Park said, “It was possible to measure health indicators including pulse and blood pressure continuously as well as pressure of body parts using our proposed soft pressure sensor. We expect it to be used in health care applications, such as the prevention and the monitoring of the pressure-driven diseases such as pressure ulcers in the near future. There will be more opportunities for future research including a whole-body pressure monitoring system related to other physical parameters.”
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Ministry of Science and ICT.
< Figure 1. The front cover image of Advanced Healthcare Materials, Volume 8, Issue 22. >
< Figure 2. Highly sensitive liquid metal-based soft pressure sensor integrated with 3D-printed microbump array. >
< Figure 3. High pressure sensitivity and reliable sensing performances of the proposed sensor and wireless heel pressure monitoring application. >
-ProfileProfessor Inkyu ParkMicro/Nano Transducers Laboratoryhttp://mintlab1.kaist.ac.kr/
Department of Mechanical EngineeringKAIST
2019.12.20 View 17571 -
 AI to Determine When to Intervene with Your Driving
(Professor Uichin Lee (left) and PhD candidate Auk Kim)
Can your AI agent judge when to talk to you while you are driving? According to a KAIST research team, their in-vehicle conservation service technology will judge when it is appropriate to contact you to ensure your safety.
Professor Uichin Lee from the Department of Industrial and Systems Engineering at KAIST and his research team have developed AI technology that automatically detects safe moments for AI agents to provide conversation services to drivers.
Their research focuses on solving the potential problems of distraction created by in-vehicle conversation services. If an AI agent talks to a driver at an inopportune moment, such as while making a turn, a car accident will be more likely to occur.
In-vehicle conversation services need to be convenient as well as safe. However, the cognitive burden of multitasking negatively influences the quality of the service. Users tend to be more distracted during certain traffic conditions. To address this long-standing challenge of the in-vehicle conversation services, the team introduced a composite cognitive model that considers both safe driving and auditory-verbal service performance and used a machine-learning model for all collected data.
The combination of these individual measures is able to determine the appropriate moments for conversation and most appropriate types of conversational services. For instance, in the case of delivering simple-context information, such as a weather forecast, driver safety alone would be the most appropriate consideration. Meanwhile, when delivering information that requires a driver response, such as a “Yes” or “No,” the combination of driver safety and auditory-verbal performance should be considered.
The research team developed a prototype of an in-vehicle conversation service based on a navigation app that can be used in real driving environments. The app was also connected to the vehicle to collect in-vehicle OBD-II/CAN data, such as the steering wheel angle and brake pedal position, and mobility and environmental data such as the distance between successive cars and traffic flow.
Using pseudo-conversation services, the research team collected a real-world driving dataset consisting of 1,388 interactions and sensor data from 29 drivers who interacted with AI conversational agents. Machine learning analysis based on the dataset demonstrated that the opportune moments for driver interruption could be correctly inferred with 87% accuracy.
The safety enhancement technology developed by the team is expected to minimize driver distractions caused by in-vehicle conversation services. This technology can be directly applied to current in-vehicle systems that provide conversation services. It can also be extended and applied to the real-time detection of driver distraction problems caused by the use of a smartphone while driving.
Professor Lee said, “In the near future, cars will proactively deliver various in-vehicle conversation services. This technology will certainly help vehicles interact with their drivers safely as it can fairly accurately determine when to provide conversation services using only basic sensor data generated by cars.”
The researchers presented their findings at the ACM International Joint Conference on Pervasive and Ubiquitous Computing (Ubicomp’19) in London, UK. This research was supported in part by Hyundai NGV and by the Next-Generation Information Computing Development Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT.
(Figure: Visual description of safe enhancement technology for in-vehicle conversation services)
2019.11.13 View 21023
AI to Determine When to Intervene with Your Driving
(Professor Uichin Lee (left) and PhD candidate Auk Kim)
Can your AI agent judge when to talk to you while you are driving? According to a KAIST research team, their in-vehicle conservation service technology will judge when it is appropriate to contact you to ensure your safety.
Professor Uichin Lee from the Department of Industrial and Systems Engineering at KAIST and his research team have developed AI technology that automatically detects safe moments for AI agents to provide conversation services to drivers.
Their research focuses on solving the potential problems of distraction created by in-vehicle conversation services. If an AI agent talks to a driver at an inopportune moment, such as while making a turn, a car accident will be more likely to occur.
In-vehicle conversation services need to be convenient as well as safe. However, the cognitive burden of multitasking negatively influences the quality of the service. Users tend to be more distracted during certain traffic conditions. To address this long-standing challenge of the in-vehicle conversation services, the team introduced a composite cognitive model that considers both safe driving and auditory-verbal service performance and used a machine-learning model for all collected data.
The combination of these individual measures is able to determine the appropriate moments for conversation and most appropriate types of conversational services. For instance, in the case of delivering simple-context information, such as a weather forecast, driver safety alone would be the most appropriate consideration. Meanwhile, when delivering information that requires a driver response, such as a “Yes” or “No,” the combination of driver safety and auditory-verbal performance should be considered.
The research team developed a prototype of an in-vehicle conversation service based on a navigation app that can be used in real driving environments. The app was also connected to the vehicle to collect in-vehicle OBD-II/CAN data, such as the steering wheel angle and brake pedal position, and mobility and environmental data such as the distance between successive cars and traffic flow.
Using pseudo-conversation services, the research team collected a real-world driving dataset consisting of 1,388 interactions and sensor data from 29 drivers who interacted with AI conversational agents. Machine learning analysis based on the dataset demonstrated that the opportune moments for driver interruption could be correctly inferred with 87% accuracy.
The safety enhancement technology developed by the team is expected to minimize driver distractions caused by in-vehicle conversation services. This technology can be directly applied to current in-vehicle systems that provide conversation services. It can also be extended and applied to the real-time detection of driver distraction problems caused by the use of a smartphone while driving.
Professor Lee said, “In the near future, cars will proactively deliver various in-vehicle conversation services. This technology will certainly help vehicles interact with their drivers safely as it can fairly accurately determine when to provide conversation services using only basic sensor data generated by cars.”
The researchers presented their findings at the ACM International Joint Conference on Pervasive and Ubiquitous Computing (Ubicomp’19) in London, UK. This research was supported in part by Hyundai NGV and by the Next-Generation Information Computing Development Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT.
(Figure: Visual description of safe enhancement technology for in-vehicle conversation services)
2019.11.13 View 21023 -
 Ultrafast Quantum Motion in a Nanoscale Trap Detected
< Professor Heung-Sun Sim (left) and Co-author Dr. Sungguen Ryu (right) >
KAIST researchers have reported the detection of a picosecond electron motion in a silicon transistor. This study has presented a new protocol for measuring ultrafast electronic dynamics in an effective time-resolved fashion of picosecond resolution. The detection was made in collaboration with Nippon Telegraph and Telephone Corp. (NTT) in Japan and National Physical Laboratory (NPL) in the UK and is the first report to the best of our knowledge.
When an electron is captured in a nanoscale trap in solids, its quantum mechanical wave function can exhibit spatial oscillation at sub-terahertz frequencies. Time-resolved detection of such picosecond dynamics of quantum waves is important, as the detection provides a way of understanding the quantum behavior of electrons in nano-electronics. It also applies to quantum information technologies such as the ultrafast quantum-bit operation of quantum computing and high-sensitivity electromagnetic-field sensing. However, detecting picosecond dynamics has been a challenge since the sub-terahertz scale is far beyond the latest bandwidth measurement tools.
A KAIST team led by Professor Heung-Sun Sim developed a theory of ultrafast electron dynamics in a nanoscale trap, and proposed a scheme for detecting the dynamics, which utilizes a quantum-mechanical resonant state formed beside the trap. The coupling between the electron dynamics and the resonant state is switched on and off at a picosecond so that information on the dynamics is read out on the electric current being generated when the coupling is switched on.
NTT realized, together with NPL, the detection scheme and applied it to electron motions in a nanoscale trap formed in a silicon transistor. A single electron was captured in the trap by controlling electrostatic gates, and a resonant state was formed in the potential barrier of the trap.
The switching on and off of the coupling between the electron and the resonant state was achieved by aligning the resonance energy with the energy of the electron within a picosecond. An electric current from the trap through the resonant state to an electrode was measured at only a few Kelvin degrees, unveiling the spatial quantum-coherent oscillation of the electron with 250 GHz frequency inside the trap.
Professor Sim said, “This work suggests a scheme of detecting picosecond electron motions in submicron scales by utilizing quantum resonance. It will be useful in dynamical control of quantum mechanical electron waves for various purposes in nano-electronics, quantum sensing, and quantum information”.
This work was published online at Nature Nanotechnology on November 4. It was partly supported by the Korea National Research Foundation through the SRC Center for Quantum Coherence in Condensed Matter. For more on the NTT news release this article, please visit https://www.ntt.co.jp/news2019/1911e/191105a.html
-ProfileProfessor Heung-Sun Sim
Department of PhysicsDirector, SRC Center for Quantum Coherence in Condensed Matterhttps://qet.kaist.ac.kr KAIST
-Publication:Gento Yamahata, Sungguen Ryu, Nathan Johnson, H.-S. Sim, Akira Fujiwara, and Masaya Kataoka. 2019. Picosecond coherent electron motion in a silicon single-electron source. Nature Nanotechnology (Online Publication). 6 pages. https://doi.org/10.1038/s41565-019-0563-2
2019.11.05 View 20952
Ultrafast Quantum Motion in a Nanoscale Trap Detected
< Professor Heung-Sun Sim (left) and Co-author Dr. Sungguen Ryu (right) >
KAIST researchers have reported the detection of a picosecond electron motion in a silicon transistor. This study has presented a new protocol for measuring ultrafast electronic dynamics in an effective time-resolved fashion of picosecond resolution. The detection was made in collaboration with Nippon Telegraph and Telephone Corp. (NTT) in Japan and National Physical Laboratory (NPL) in the UK and is the first report to the best of our knowledge.
When an electron is captured in a nanoscale trap in solids, its quantum mechanical wave function can exhibit spatial oscillation at sub-terahertz frequencies. Time-resolved detection of such picosecond dynamics of quantum waves is important, as the detection provides a way of understanding the quantum behavior of electrons in nano-electronics. It also applies to quantum information technologies such as the ultrafast quantum-bit operation of quantum computing and high-sensitivity electromagnetic-field sensing. However, detecting picosecond dynamics has been a challenge since the sub-terahertz scale is far beyond the latest bandwidth measurement tools.
A KAIST team led by Professor Heung-Sun Sim developed a theory of ultrafast electron dynamics in a nanoscale trap, and proposed a scheme for detecting the dynamics, which utilizes a quantum-mechanical resonant state formed beside the trap. The coupling between the electron dynamics and the resonant state is switched on and off at a picosecond so that information on the dynamics is read out on the electric current being generated when the coupling is switched on.
NTT realized, together with NPL, the detection scheme and applied it to electron motions in a nanoscale trap formed in a silicon transistor. A single electron was captured in the trap by controlling electrostatic gates, and a resonant state was formed in the potential barrier of the trap.
The switching on and off of the coupling between the electron and the resonant state was achieved by aligning the resonance energy with the energy of the electron within a picosecond. An electric current from the trap through the resonant state to an electrode was measured at only a few Kelvin degrees, unveiling the spatial quantum-coherent oscillation of the electron with 250 GHz frequency inside the trap.
Professor Sim said, “This work suggests a scheme of detecting picosecond electron motions in submicron scales by utilizing quantum resonance. It will be useful in dynamical control of quantum mechanical electron waves for various purposes in nano-electronics, quantum sensing, and quantum information”.
This work was published online at Nature Nanotechnology on November 4. It was partly supported by the Korea National Research Foundation through the SRC Center for Quantum Coherence in Condensed Matter. For more on the NTT news release this article, please visit https://www.ntt.co.jp/news2019/1911e/191105a.html
-ProfileProfessor Heung-Sun Sim
Department of PhysicsDirector, SRC Center for Quantum Coherence in Condensed Matterhttps://qet.kaist.ac.kr KAIST
-Publication:Gento Yamahata, Sungguen Ryu, Nathan Johnson, H.-S. Sim, Akira Fujiwara, and Masaya Kataoka. 2019. Picosecond coherent electron motion in a silicon single-electron source. Nature Nanotechnology (Online Publication). 6 pages. https://doi.org/10.1038/s41565-019-0563-2
2019.11.05 View 20952 -
 A Mathematical Model Reveals Long-Distance Cell Communication Mechanism
How can tens of thousands of people in a large football stadium all clap together with the same beat even though they can only hear the people near them clapping?
A combination of a partial differential equation and a synthetic circuit in microbes answers this question. An interdisciplinary collaborative team of Professor Jae Kyoung Kim at KAIST, Professor Krešimir Josić at the University of Houston, and Professor Matt Bennett at Rice University has identified how a large community can communicate with each other almost simultaneously even with very short distance signaling. The research was reported at Nature Chemical Biology.
Cells often communicate using signaling molecules, which can travel only a short distance. Nevertheless, the cells can also communicate over large distances to spur collective action. The team revealed a cell communication mechanism that quickly forms a network of local interactions to spur collective action, even in large communities.
The research team used an engineered transcriptional circuit of combined positive and negative feedback loops in E. coli, which can periodically release two types of signaling molecules: activator and repressor. As the signaling molecules travel over a short distance, cells can only talk to their nearest neighbors. However, cell communities synchronize oscillatory gene expression in spatially extended systems as long as the transcriptional circuit contains a positive feedback loop for the activator.
Professor Kim said that analyzing and understanding such high-dimensional dynamics was extremely difficult. He explained, “That’s why we used high-dimensional partial differential equation to describe the system based on the interactions among various types of molecules.” Surprisingly, the mathematical model accurately simulates the synthesis of the signaling molecules in the cell and their spatial diffusion throughout the chamber and their effect on neighboring cells.
The team simplified the high-dimensional system into a one-dimensional orbit, noting that the system repeats periodically. This allowed them to discover that cells can make one voice when they lowered their own voice and listened to the others. “It turns out the positive feedback loop reduces the distance between moving points and finally makes them move all together. That’s why you clap louder when you hear applause from nearby neighbors and everyone eventually claps together at almost the same time,” said Professor Kim.
Professor Kim added, “Math is a powerful as it simplifies complex thing so that we can find an essential underlying property. This finding would not have been possible without the simplification of complex systems using mathematics."
The National Institutes of Health, the National Science Foundation, the Robert A. Welch Foundation, the Hamill Foundation, the National Research Foundation of Korea, and the T.J. Park Science Fellowship of POSCO supported the research.
(Figure: Complex molecular interactions among microbial consortia is simplified as interactions among points on a limit cycle (right).)
2019.10.15 View 31278
A Mathematical Model Reveals Long-Distance Cell Communication Mechanism
How can tens of thousands of people in a large football stadium all clap together with the same beat even though they can only hear the people near them clapping?
A combination of a partial differential equation and a synthetic circuit in microbes answers this question. An interdisciplinary collaborative team of Professor Jae Kyoung Kim at KAIST, Professor Krešimir Josić at the University of Houston, and Professor Matt Bennett at Rice University has identified how a large community can communicate with each other almost simultaneously even with very short distance signaling. The research was reported at Nature Chemical Biology.
Cells often communicate using signaling molecules, which can travel only a short distance. Nevertheless, the cells can also communicate over large distances to spur collective action. The team revealed a cell communication mechanism that quickly forms a network of local interactions to spur collective action, even in large communities.
The research team used an engineered transcriptional circuit of combined positive and negative feedback loops in E. coli, which can periodically release two types of signaling molecules: activator and repressor. As the signaling molecules travel over a short distance, cells can only talk to their nearest neighbors. However, cell communities synchronize oscillatory gene expression in spatially extended systems as long as the transcriptional circuit contains a positive feedback loop for the activator.
Professor Kim said that analyzing and understanding such high-dimensional dynamics was extremely difficult. He explained, “That’s why we used high-dimensional partial differential equation to describe the system based on the interactions among various types of molecules.” Surprisingly, the mathematical model accurately simulates the synthesis of the signaling molecules in the cell and their spatial diffusion throughout the chamber and their effect on neighboring cells.
The team simplified the high-dimensional system into a one-dimensional orbit, noting that the system repeats periodically. This allowed them to discover that cells can make one voice when they lowered their own voice and listened to the others. “It turns out the positive feedback loop reduces the distance between moving points and finally makes them move all together. That’s why you clap louder when you hear applause from nearby neighbors and everyone eventually claps together at almost the same time,” said Professor Kim.
Professor Kim added, “Math is a powerful as it simplifies complex thing so that we can find an essential underlying property. This finding would not have been possible without the simplification of complex systems using mathematics."
The National Institutes of Health, the National Science Foundation, the Robert A. Welch Foundation, the Hamill Foundation, the National Research Foundation of Korea, and the T.J. Park Science Fellowship of POSCO supported the research.
(Figure: Complex molecular interactions among microbial consortia is simplified as interactions among points on a limit cycle (right).)
2019.10.15 View 31278 -
 Professor Byong-Guk Park Named Scientist of October
< Professor Byong-Guk Park >
Professor Byong-Guk Park from the Department of Materials Science and Engineering was selected as the ‘Scientist of the Month’ for October 2019 by the Ministry of Science and ICT and the National Research Foundation of Korea. Professor Park was recognized for his contributions to the advancement of spin-orbit torque (SOT)-based magnetic random access memory (MRAM) technology. He received 10 million KRW in prize money.
A next-generation, non-volatile memory device MRAM consists of thin magnetic films. It can be applied in “logic-in-memory” devices, in which logic and memory functionalities coexist, thus drastically improving the performance of complementary metal-oxide semiconductor (CMOS) processors. Conventional MRAM technology is limited in its ability to increase the operation speed of a memory device while maintaining a high density.
Professor Park tackled this challenge by introducing a new material, antiferromagnet (IrMn), that generates a sizable amount of SOT as well as an exchange-bias field, which makes successful data writing possible without an external magnetic field. This research outcome paved the way for the development of MRAM, which has a simple device structure but features high speeds and density.
Professor Park said, “I feel rewarded to have forwarded the feasibility and applicability of MRAM. I will continue devoting myself to studying further on the development of new materials that can help enhance the performance of memory devices."
(END)
2019.10.10 View 12751
Professor Byong-Guk Park Named Scientist of October
< Professor Byong-Guk Park >
Professor Byong-Guk Park from the Department of Materials Science and Engineering was selected as the ‘Scientist of the Month’ for October 2019 by the Ministry of Science and ICT and the National Research Foundation of Korea. Professor Park was recognized for his contributions to the advancement of spin-orbit torque (SOT)-based magnetic random access memory (MRAM) technology. He received 10 million KRW in prize money.
A next-generation, non-volatile memory device MRAM consists of thin magnetic films. It can be applied in “logic-in-memory” devices, in which logic and memory functionalities coexist, thus drastically improving the performance of complementary metal-oxide semiconductor (CMOS) processors. Conventional MRAM technology is limited in its ability to increase the operation speed of a memory device while maintaining a high density.
Professor Park tackled this challenge by introducing a new material, antiferromagnet (IrMn), that generates a sizable amount of SOT as well as an exchange-bias field, which makes successful data writing possible without an external magnetic field. This research outcome paved the way for the development of MRAM, which has a simple device structure but features high speeds and density.
Professor Park said, “I feel rewarded to have forwarded the feasibility and applicability of MRAM. I will continue devoting myself to studying further on the development of new materials that can help enhance the performance of memory devices."
(END)
2019.10.10 View 12751 -
 Researchers Describe a Mechanism Inducing Self-Killing of Cancer Cells
(Professor Kim (left) and lead author Lee)
Researchers have described a new mechanism which induces the self-killing of cancer cells by perturbing ion homeostasis. A research team from the Department of Biochemical Engineering has developed helical polypeptide potassium ionophores that lead to the onset of programmed cell death. The ionophores increase the active oxygen concentration to stress endoplasmic reticulum to the point of cellular death.
The electrochemical gradient between extracellular and intracellular conditions plays an important role in cell growth and metabolism. When a cell’s ion homeostasis is disturbed, critical functions accelerating the activation of apoptosis are inhibited in the cell.
Although ionophores have been intensively used as an ion homeostasis disturber, the mechanisms of cell death have been unclear and the bio-applicability has been limited. In the study featured at Advanced Science, the team presented an alpha helical peptide-based anticancer agent that is capable of transporting potassium ions with water solubility. The cationic, hydrophilic, and potassium ionic groups were combined at the end of the peptide side chain to provide both ion transport and hydrophilic properties.
These peptide-based ionophores reduce the intracellular potassium concentration and at the same time increase the intracellular calcium concentration. Increased intracellular calcium concentrations produce intracellular reactive oxygen species, causing endoplasmic reticulum stress, and ultimately leading to apoptosis.
Anticancer effects were evaluated using tumor-bearing mice to confirm the therapeutic effect, even in animal models. It was found that tumor growth was strongly inhibited by endoplasmic stress-mediated apoptosis.
Lead author Dr. Dae-Yong Lee said, “A peptide-based ionophore is more effective than conventional chemotherapeutic agents because it induces apoptosis via elevated reactive oxygen species levels. Professor Yeu-Chun Kim said he expects this new mechanism to be widely used as a new chemotherapeutic strategy. This research was funded by the National Research Foundation.
2019.08.28 View 23048
Researchers Describe a Mechanism Inducing Self-Killing of Cancer Cells
(Professor Kim (left) and lead author Lee)
Researchers have described a new mechanism which induces the self-killing of cancer cells by perturbing ion homeostasis. A research team from the Department of Biochemical Engineering has developed helical polypeptide potassium ionophores that lead to the onset of programmed cell death. The ionophores increase the active oxygen concentration to stress endoplasmic reticulum to the point of cellular death.
The electrochemical gradient between extracellular and intracellular conditions plays an important role in cell growth and metabolism. When a cell’s ion homeostasis is disturbed, critical functions accelerating the activation of apoptosis are inhibited in the cell.
Although ionophores have been intensively used as an ion homeostasis disturber, the mechanisms of cell death have been unclear and the bio-applicability has been limited. In the study featured at Advanced Science, the team presented an alpha helical peptide-based anticancer agent that is capable of transporting potassium ions with water solubility. The cationic, hydrophilic, and potassium ionic groups were combined at the end of the peptide side chain to provide both ion transport and hydrophilic properties.
These peptide-based ionophores reduce the intracellular potassium concentration and at the same time increase the intracellular calcium concentration. Increased intracellular calcium concentrations produce intracellular reactive oxygen species, causing endoplasmic reticulum stress, and ultimately leading to apoptosis.
Anticancer effects were evaluated using tumor-bearing mice to confirm the therapeutic effect, even in animal models. It was found that tumor growth was strongly inhibited by endoplasmic stress-mediated apoptosis.
Lead author Dr. Dae-Yong Lee said, “A peptide-based ionophore is more effective than conventional chemotherapeutic agents because it induces apoptosis via elevated reactive oxygen species levels. Professor Yeu-Chun Kim said he expects this new mechanism to be widely used as a new chemotherapeutic strategy. This research was funded by the National Research Foundation.
2019.08.28 View 23048 -
 Accurate Detection of Low-Level Somatic Mutation in Intractable Epilepsy
KAIST medical scientists have developed an advanced method for perfectly detecting low-level somatic mutation in patients with intractable epilepsy. Their study showed that deep sequencing replicates of major focal epilepsy genes accurately and efficiently identified low-level somatic mutations in intractable epilepsy.
According to the study, their diagnostic method could increase the accuracy up to 100%, unlike the conventional sequencing analysis, which stands at about 30% accuracy. This work was published in Acta Neuropathologica.
Epilepsy is a neurological disorder common in children. Approximately one third of child patients are diagnosed with intractable epilepsy despite adequate anti-epileptic medication treatment.
Somatic mutations in mTOR pathway genes, SLC35A2, and BRAF are the major genetic causes of intractable epilepsies. A clinical trial to target Focal Cortical Dysplasia type II (FCDII), the mTOR inhibitor is underway at Severance Hospital, their collaborator in Seoul, Korea. However, it is difficult to detect such somatic mutations causing intractable epilepsy because their mutational burden is less than 5%, which is similar to the level of sequencing artifacts. In the clinical field, this has remained a standing challenge for the genetic diagnosis of somatic mutations in intractable epilepsy.
Professor Jeong Ho Lee’s team at the Graduate School of Medical Science and Engineering analyzed paired brain and peripheral tissues from 232 intractable epilepsy patients with various brain pathologies at Severance Hospital using deep sequencing and extracted the major focal epilepsy genes.
They narrowed down target genes to eight major focal epilepsy genes, eliminating almost all of the false positive calls using deep targeted sequencing. As a result, the advanced method robustly increased the accuracy and enabled them to detect low-level somatic mutations in unmatched Formalin Fixed Paraffin Embedded (FFPE) brain samples, the most clinically relevant samples.
Professor Lee conducted this study in collaboration with Professor Dong Suk Kim and Hoon-Chul Kang at Severance Hospital of Yonsei University. He said, “This advanced method of genetic analysis will improve overall patient care by providing more comprehensive genetic counseling and informing decisions on alternative treatments.”
Professor Lee has investigated low-level somatic mutations arising in the brain for a decade. He is developing innovative diagnostics and therapeutics for untreatable brain disorders including intractable epilepsy and glioblastoma at a tech-startup called SoVarGen. “All of the technologies we used during the research were transferred to the company. This research gave us very good momentum to reach the next phase of our startup,” he remarked.
The work was supported by grants from the Suh Kyungbae Foundation, a National Research Foundation of Korea grant funded by the Ministry of Science and ICT, the Korean Health Technology R&D Project from the Ministry of Health & Welfare, and the Netherlands Organization for Health Research and Development.
(Figure: Landscape of somatic and germline mutations identified in intractable epilepsy patients. a Signaling pathways for all of the mutated genes identified in this study. Bold: somatic mutation, Regular: germline mutation. b The distribution of variant allelic frequencies (VAFs) of identified somatic mutations. c The detecting rate and types of identified mutations according to histopathology. Yellow: somatic mutations, green: two-hit mutations, grey: germline mutations.)
2019.08.14 View 31298
Accurate Detection of Low-Level Somatic Mutation in Intractable Epilepsy
KAIST medical scientists have developed an advanced method for perfectly detecting low-level somatic mutation in patients with intractable epilepsy. Their study showed that deep sequencing replicates of major focal epilepsy genes accurately and efficiently identified low-level somatic mutations in intractable epilepsy.
According to the study, their diagnostic method could increase the accuracy up to 100%, unlike the conventional sequencing analysis, which stands at about 30% accuracy. This work was published in Acta Neuropathologica.
Epilepsy is a neurological disorder common in children. Approximately one third of child patients are diagnosed with intractable epilepsy despite adequate anti-epileptic medication treatment.
Somatic mutations in mTOR pathway genes, SLC35A2, and BRAF are the major genetic causes of intractable epilepsies. A clinical trial to target Focal Cortical Dysplasia type II (FCDII), the mTOR inhibitor is underway at Severance Hospital, their collaborator in Seoul, Korea. However, it is difficult to detect such somatic mutations causing intractable epilepsy because their mutational burden is less than 5%, which is similar to the level of sequencing artifacts. In the clinical field, this has remained a standing challenge for the genetic diagnosis of somatic mutations in intractable epilepsy.
Professor Jeong Ho Lee’s team at the Graduate School of Medical Science and Engineering analyzed paired brain and peripheral tissues from 232 intractable epilepsy patients with various brain pathologies at Severance Hospital using deep sequencing and extracted the major focal epilepsy genes.
They narrowed down target genes to eight major focal epilepsy genes, eliminating almost all of the false positive calls using deep targeted sequencing. As a result, the advanced method robustly increased the accuracy and enabled them to detect low-level somatic mutations in unmatched Formalin Fixed Paraffin Embedded (FFPE) brain samples, the most clinically relevant samples.
Professor Lee conducted this study in collaboration with Professor Dong Suk Kim and Hoon-Chul Kang at Severance Hospital of Yonsei University. He said, “This advanced method of genetic analysis will improve overall patient care by providing more comprehensive genetic counseling and informing decisions on alternative treatments.”
Professor Lee has investigated low-level somatic mutations arising in the brain for a decade. He is developing innovative diagnostics and therapeutics for untreatable brain disorders including intractable epilepsy and glioblastoma at a tech-startup called SoVarGen. “All of the technologies we used during the research were transferred to the company. This research gave us very good momentum to reach the next phase of our startup,” he remarked.
The work was supported by grants from the Suh Kyungbae Foundation, a National Research Foundation of Korea grant funded by the Ministry of Science and ICT, the Korean Health Technology R&D Project from the Ministry of Health & Welfare, and the Netherlands Organization for Health Research and Development.
(Figure: Landscape of somatic and germline mutations identified in intractable epilepsy patients. a Signaling pathways for all of the mutated genes identified in this study. Bold: somatic mutation, Regular: germline mutation. b The distribution of variant allelic frequencies (VAFs) of identified somatic mutations. c The detecting rate and types of identified mutations according to histopathology. Yellow: somatic mutations, green: two-hit mutations, grey: germline mutations.)
2019.08.14 View 31298 -
 Manipulating Brain Cells by Smartphone
Researchers have developed a soft neural implant that can be wirelessly controlled using a smartphone. It is the first wireless neural device capable of indefinitely delivering multiple drugs and multiple colour lights, which neuroscientists believe can speed up efforts to uncover brain diseases such as Parkinson’s, Alzheimer’s, addiction, depression, and pain.
A team under Professor Jae-Woong Jeong from the School of Electrical Engineering at KAIST and his collaborators have invented a device that can control neural circuits using a tiny brain implant controlled by a smartphone. The device, using Lego-like replaceable drug cartridges and powerful, low-energy Bluetooth, can target specific neurons of interest using drugs and light for prolonged periods. This study was published in Nature Biomedical Engineering.
“This novel device is the fruit of advanced electronics design and powerful micro and nanoscale engineering,” explained Professor Jeong. “We are interested in further developing this technology to make a brain implant for clinical applications.”
This technology significantly overshadows the conventional methods used by neuroscientists, which usually involve rigid metal tubes and optical fibers to deliver drugs and light. Apart from limiting the subject’s movement due to bulky equipment, their relatively rigid structure causes lesions in soft brain tissue over time, therefore making them not suitable for long-term implantation. Although some efforts have been made to partly mitigate adverse tissue response by incorporating soft probes and wireless platforms, the previous solutions were limited by their inability to deliver drugs for long periods of time as well as their bulky and complex control setups.
To achieve chronic wireless drug delivery, scientists had to solve the critical challenge of the exhaustion and evaporation of drugs. To combat this, the researchers invented a neural device with a replaceable drug cartridge, which could allow neuroscientists to study the same brain circuits for several months without worrying about running out of drugs.
These ‘plug-n-play’ drug cartridges were assembled into a brain implant for mice with a soft and ultrathin probe (with the thickness of a human hair), which consisted of microfluidic channels and tiny LEDs (smaller than a grain of salt), for unlimited drug doses and light delivery.
Controlled with an elegant and simple user interface on a smartphone, neuroscientists can easily trigger any specific combination or precise sequencing of light and drug delivery in any implanted target animal without the need to be physically inside the laboratory. Using these wireless neural devices, researchers can also easily setup fully automated animal studies where the behaviour of one animal could affect other animals by triggering light and/or drug delivery.
“The wireless neural device enables chronic chemical and optical neuromodulation that has never been achieved before,” said lead author Raza Qazi, a researcher with KAIST and the University of Colorado Boulder.
This work was supported by grants from the National Research Foundation of Korea, US National Institute of Health, National Institute on Drug Abuse, and Mallinckrodt Professorship.
(A neural implant with replaceable drug cartridges and Bluetooth low-energy can target specific neurons .)
(Micro LED controlling using smartphone application)
2019.08.07 View 35266
Manipulating Brain Cells by Smartphone
Researchers have developed a soft neural implant that can be wirelessly controlled using a smartphone. It is the first wireless neural device capable of indefinitely delivering multiple drugs and multiple colour lights, which neuroscientists believe can speed up efforts to uncover brain diseases such as Parkinson’s, Alzheimer’s, addiction, depression, and pain.
A team under Professor Jae-Woong Jeong from the School of Electrical Engineering at KAIST and his collaborators have invented a device that can control neural circuits using a tiny brain implant controlled by a smartphone. The device, using Lego-like replaceable drug cartridges and powerful, low-energy Bluetooth, can target specific neurons of interest using drugs and light for prolonged periods. This study was published in Nature Biomedical Engineering.
“This novel device is the fruit of advanced electronics design and powerful micro and nanoscale engineering,” explained Professor Jeong. “We are interested in further developing this technology to make a brain implant for clinical applications.”
This technology significantly overshadows the conventional methods used by neuroscientists, which usually involve rigid metal tubes and optical fibers to deliver drugs and light. Apart from limiting the subject’s movement due to bulky equipment, their relatively rigid structure causes lesions in soft brain tissue over time, therefore making them not suitable for long-term implantation. Although some efforts have been made to partly mitigate adverse tissue response by incorporating soft probes and wireless platforms, the previous solutions were limited by their inability to deliver drugs for long periods of time as well as their bulky and complex control setups.
To achieve chronic wireless drug delivery, scientists had to solve the critical challenge of the exhaustion and evaporation of drugs. To combat this, the researchers invented a neural device with a replaceable drug cartridge, which could allow neuroscientists to study the same brain circuits for several months without worrying about running out of drugs.
These ‘plug-n-play’ drug cartridges were assembled into a brain implant for mice with a soft and ultrathin probe (with the thickness of a human hair), which consisted of microfluidic channels and tiny LEDs (smaller than a grain of salt), for unlimited drug doses and light delivery.
Controlled with an elegant and simple user interface on a smartphone, neuroscientists can easily trigger any specific combination or precise sequencing of light and drug delivery in any implanted target animal without the need to be physically inside the laboratory. Using these wireless neural devices, researchers can also easily setup fully automated animal studies where the behaviour of one animal could affect other animals by triggering light and/or drug delivery.
“The wireless neural device enables chronic chemical and optical neuromodulation that has never been achieved before,” said lead author Raza Qazi, a researcher with KAIST and the University of Colorado Boulder.
This work was supported by grants from the National Research Foundation of Korea, US National Institute of Health, National Institute on Drug Abuse, and Mallinckrodt Professorship.
(A neural implant with replaceable drug cartridges and Bluetooth low-energy can target specific neurons .)
(Micro LED controlling using smartphone application)
2019.08.07 View 35266 -
 Synthesizing Single-Crystalline Hexagonal Graphene Quantum Dots
(Figure: Uniformly ordered single-crystalline graphene quantum dots of various sizes synthesized through solution chemistry.)
A KAIST team has designed a novel strategy for synthesizing single-crystalline graphene quantum dots, which emit stable blue light. The research team confirmed that a display made of their synthesized graphene quantum dots successfully emitted blue light with stable electric pressure, reportedly resolving the long-standing challenges of blue light emission in manufactured displays. The study, led by Professor O Ok Park in the Department of Chemical and Biological Engineering, was featured online in Nano Letters on July 5.
Graphene has gained increased attention as a next-generation material for its heat and electrical conductivity as well as its transparency. However, single and multi-layered graphene have characteristics of a conductor so that it is difficult to apply into semiconductor. Only when downsized to the nanoscale, semiconductor’s distinct feature of bandgap will be exhibited to emit the light in the graphene. This illuminating featuring of dot is referred to as a graphene quantum dot.
Conventionally, single-crystalline graphene has been fabricated by chemical vapor deposition (CVD) on copper or nickel thin films, or by peeling graphite physically and chemically. However, graphene made via chemical vapor deposition is mainly used for large-surface transparent electrodes. Meanwhile, graphene made by chemical and physical peeling carries uneven size defects.
The research team explained that their graphene quantum dots exhibited a very stable single-phase reaction when they mixed amine and acetic acid with an aqueous solution of glucose. Then, they synthesized single-crystalline graphene quantum dots from the self-assembly of the reaction intermediate. In the course of fabrication, the team developed a new separation method at a low-temperature precipitation, which led to successfully creating a homogeneous nucleation of graphene quantum dots via a single-phase reaction.
Professor Park and his colleagues have developed solution phase synthesis technology that allows for the creation of the desired crystal size for single nanocrystals down to 100 nano meters. It is reportedly the first synthesis of the homogeneous nucleation of graphene through a single-phase reaction.
Professor Park said, "This solution method will significantly contribute to the grafting of graphene in various fields. The application of this new graphene will expand the scope of its applications such as for flexible displays and varistors.”
This research was a joint project with a team from Korea University under Professor Sang Hyuk Im from the Department of Chemical and Biological Engineering, and was supported by the National Research Foundation of Korea, the Nano-Material Technology Development Program from the Electronics and Telecommunications Research Institute (ETRI), KAIST EEWS, and the BK21+ project from the Korean government.
2019.08.02 View 36817
Synthesizing Single-Crystalline Hexagonal Graphene Quantum Dots
(Figure: Uniformly ordered single-crystalline graphene quantum dots of various sizes synthesized through solution chemistry.)
A KAIST team has designed a novel strategy for synthesizing single-crystalline graphene quantum dots, which emit stable blue light. The research team confirmed that a display made of their synthesized graphene quantum dots successfully emitted blue light with stable electric pressure, reportedly resolving the long-standing challenges of blue light emission in manufactured displays. The study, led by Professor O Ok Park in the Department of Chemical and Biological Engineering, was featured online in Nano Letters on July 5.
Graphene has gained increased attention as a next-generation material for its heat and electrical conductivity as well as its transparency. However, single and multi-layered graphene have characteristics of a conductor so that it is difficult to apply into semiconductor. Only when downsized to the nanoscale, semiconductor’s distinct feature of bandgap will be exhibited to emit the light in the graphene. This illuminating featuring of dot is referred to as a graphene quantum dot.
Conventionally, single-crystalline graphene has been fabricated by chemical vapor deposition (CVD) on copper or nickel thin films, or by peeling graphite physically and chemically. However, graphene made via chemical vapor deposition is mainly used for large-surface transparent electrodes. Meanwhile, graphene made by chemical and physical peeling carries uneven size defects.
The research team explained that their graphene quantum dots exhibited a very stable single-phase reaction when they mixed amine and acetic acid with an aqueous solution of glucose. Then, they synthesized single-crystalline graphene quantum dots from the self-assembly of the reaction intermediate. In the course of fabrication, the team developed a new separation method at a low-temperature precipitation, which led to successfully creating a homogeneous nucleation of graphene quantum dots via a single-phase reaction.
Professor Park and his colleagues have developed solution phase synthesis technology that allows for the creation of the desired crystal size for single nanocrystals down to 100 nano meters. It is reportedly the first synthesis of the homogeneous nucleation of graphene through a single-phase reaction.
Professor Park said, "This solution method will significantly contribute to the grafting of graphene in various fields. The application of this new graphene will expand the scope of its applications such as for flexible displays and varistors.”
This research was a joint project with a team from Korea University under Professor Sang Hyuk Im from the Department of Chemical and Biological Engineering, and was supported by the National Research Foundation of Korea, the Nano-Material Technology Development Program from the Electronics and Telecommunications Research Institute (ETRI), KAIST EEWS, and the BK21+ project from the Korean government.
2019.08.02 View 36817 -
 Mathematical Modeling Makes a Breakthrough for a New CRSD Medication
PhD Candidate Dae Wook Kim (Left) and Professor Jae Kyoung Kim (Right)
- Systems approach reveals photosensitivity and PER2 level as determinants of clock-modulator efficacy -
Mathematicians’ new modeling has identified major sources of interspecies and inter-individual variations in the clinical efficacy of a clock-modulating drug: photosensitivity and PER2 level. This enabled precision medicine for circadian disruption.
A KAIST mathematics research team led by Professor Jae Kyoung Kim, in collaboration with Pfizer, applied a combination of mathematical modeling and simulation tools for circadian rhythms sleep disorders (CRSDs) to analyze the animal data generated by Pfizer. This study was reported in Molecular Systems Biology as the cover article on July 8.
Pharmaceutical companies have conducted extensive studies on animals to determine the candidacy of this new medication. However, the results of animal testing do not always translate to the same effects in human trials. Furthermore, even between humans, efficacy differs across individuals depending on an individual’s genetic and environmental factors, which require different treatment strategies.
To overcome these obstacles, KAIST mathematicians and their collaborators developed adaptive chronotherapeutics to identify precise dosing regimens that could restore normal circadian phase under different conditions.
A circadian rhythm is a 24-hour cycle in the physiological processes of living creatures, including humans. A biological clock in the hypothalamic suprachiasmatic nucleus in the human brain sets the time for various human behaviors such as sleep.
A disruption of the endogenous timekeeping system caused by changes in one’s life pattern leads to advanced or delayed sleep-wake cycle phase and a desynchronization between sleep-wake rhythms, resulting in CRSDs. To restore the normal timing of sleep, timing of the circadian clock could be adjusted pharmacologically.
Pfizer identified PF-670462, which can adjust the timing of circadian clock by inhibiting the core clock kinase of the circadian clock (CK1d/e). However, the efficacy of PF-670462 significantly differs between nocturnal mice and diurnal monkeys, whose sleeping times are opposite.
The research team discovered the source of such interspecies variations in drug response by performing thousands of virtual experiments using a mathematical model, which describes biochemical interactions among clock molecules and PF-670462. The result suggests that the effect of PF-670462 is reduced by light exposure in diurnal primates more than in nocturnal mice. This indicates that the strong counteracting effect of light must be considered in order to effectively regulate the circadian clock of diurnal humans using PF-670462.
Furthermore, the team also found the source of inter-patients variations in drug efficacy using virtual patients whose circadian clocks were disrupted due to various mutations. The degree of perturbation in the endogenous level of the core clock molecule PER2 affects the efficacy.
This explains why the clinical outcomes of clock-modulating drugs are highly variable and certain subtypes are unresponsive to treatment. Furthermore, this points out the limitations of current treatment strategies tailored to only the patient’s sleep and wake time but not to the molecular cause of sleep disorders.
PhD candidate Dae Wook Kim, who is the first author, said that this motivates the team to develop an adaptive chronotherapy, which identifies a personalized optimal dosing time of day by tracking the sleep-wake up time of patients via a wearable device and allows for a precision medicine approach for CRSDs.
Professor Jae Kyoung Kim said, "As a mathematician, I am excited to help enable the advancement of a new drug candidate, which can improve the lives of so many patients. I hope this result promotes more collaborations in this translational research.”
This research was supported by a Pfizer grant to KAIST (G01160179), the Human Frontiers Science Program Organization (RGY0063/2017), and a National Research Foundation (NRF) of Korea Grant (NRF-2016 RICIB 3008468 and NRF-2017-Fostering Core Leaders of the Future Basic Science Program/ Global Ph.D. Fellowship Program).
Figure 1. Interspecies and Inter-patients Variations in PF-670462 Efficacy
Figure 2. Journal Cover Page
Publication:
Dae Wook Kim, Cheng Chang, Xian Chen, Angela C Doran, Francois Gaudreault, Travis Wager, George J DeMarco, and Jae Kyoung Kim. 2019. Systems approach reveals photosensitivity and PER2 level as determinants of clock-modulator efficacy. Molecular Systems Biology. EMBO Press, Heidelberg, Germany, Vol. 15, Issue No. 7, Article, 16 pages. https://doi.org/10.15252/msb.20198838
Profile: Prof. Jae Kyoung Kim, PhD
jaekkim@kaist.ac.kr
http://mathsci.kaist.ac.kr/~jaekkim
Associate Professor
Department of Mathematical Sciences
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr Daejeon 34141, Korea
Profile: Dae Wook Kim, PhD Candidate
0308kdo@kaist.ac.kr
http://mathsci.kaist.ac.kr/~jaekkim
PhD Candidate
Department of Mathematical Sciences
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr Daejeon 34141, Korea
Profile: Dr. Cheng Chang, PhD
cheng.chang@pfizer.com
Associate Director of Clinical Pharmacology
Clinical Pharmacology, Global Product Development
Pfizer
https://www.pfizer.com/ Groton 06340, USA
(END)
2019.07.09 View 38222
Mathematical Modeling Makes a Breakthrough for a New CRSD Medication
PhD Candidate Dae Wook Kim (Left) and Professor Jae Kyoung Kim (Right)
- Systems approach reveals photosensitivity and PER2 level as determinants of clock-modulator efficacy -
Mathematicians’ new modeling has identified major sources of interspecies and inter-individual variations in the clinical efficacy of a clock-modulating drug: photosensitivity and PER2 level. This enabled precision medicine for circadian disruption.
A KAIST mathematics research team led by Professor Jae Kyoung Kim, in collaboration with Pfizer, applied a combination of mathematical modeling and simulation tools for circadian rhythms sleep disorders (CRSDs) to analyze the animal data generated by Pfizer. This study was reported in Molecular Systems Biology as the cover article on July 8.
Pharmaceutical companies have conducted extensive studies on animals to determine the candidacy of this new medication. However, the results of animal testing do not always translate to the same effects in human trials. Furthermore, even between humans, efficacy differs across individuals depending on an individual’s genetic and environmental factors, which require different treatment strategies.
To overcome these obstacles, KAIST mathematicians and their collaborators developed adaptive chronotherapeutics to identify precise dosing regimens that could restore normal circadian phase under different conditions.
A circadian rhythm is a 24-hour cycle in the physiological processes of living creatures, including humans. A biological clock in the hypothalamic suprachiasmatic nucleus in the human brain sets the time for various human behaviors such as sleep.
A disruption of the endogenous timekeeping system caused by changes in one’s life pattern leads to advanced or delayed sleep-wake cycle phase and a desynchronization between sleep-wake rhythms, resulting in CRSDs. To restore the normal timing of sleep, timing of the circadian clock could be adjusted pharmacologically.
Pfizer identified PF-670462, which can adjust the timing of circadian clock by inhibiting the core clock kinase of the circadian clock (CK1d/e). However, the efficacy of PF-670462 significantly differs between nocturnal mice and diurnal monkeys, whose sleeping times are opposite.
The research team discovered the source of such interspecies variations in drug response by performing thousands of virtual experiments using a mathematical model, which describes biochemical interactions among clock molecules and PF-670462. The result suggests that the effect of PF-670462 is reduced by light exposure in diurnal primates more than in nocturnal mice. This indicates that the strong counteracting effect of light must be considered in order to effectively regulate the circadian clock of diurnal humans using PF-670462.
Furthermore, the team also found the source of inter-patients variations in drug efficacy using virtual patients whose circadian clocks were disrupted due to various mutations. The degree of perturbation in the endogenous level of the core clock molecule PER2 affects the efficacy.
This explains why the clinical outcomes of clock-modulating drugs are highly variable and certain subtypes are unresponsive to treatment. Furthermore, this points out the limitations of current treatment strategies tailored to only the patient’s sleep and wake time but not to the molecular cause of sleep disorders.
PhD candidate Dae Wook Kim, who is the first author, said that this motivates the team to develop an adaptive chronotherapy, which identifies a personalized optimal dosing time of day by tracking the sleep-wake up time of patients via a wearable device and allows for a precision medicine approach for CRSDs.
Professor Jae Kyoung Kim said, "As a mathematician, I am excited to help enable the advancement of a new drug candidate, which can improve the lives of so many patients. I hope this result promotes more collaborations in this translational research.”
This research was supported by a Pfizer grant to KAIST (G01160179), the Human Frontiers Science Program Organization (RGY0063/2017), and a National Research Foundation (NRF) of Korea Grant (NRF-2016 RICIB 3008468 and NRF-2017-Fostering Core Leaders of the Future Basic Science Program/ Global Ph.D. Fellowship Program).
Figure 1. Interspecies and Inter-patients Variations in PF-670462 Efficacy
Figure 2. Journal Cover Page
Publication:
Dae Wook Kim, Cheng Chang, Xian Chen, Angela C Doran, Francois Gaudreault, Travis Wager, George J DeMarco, and Jae Kyoung Kim. 2019. Systems approach reveals photosensitivity and PER2 level as determinants of clock-modulator efficacy. Molecular Systems Biology. EMBO Press, Heidelberg, Germany, Vol. 15, Issue No. 7, Article, 16 pages. https://doi.org/10.15252/msb.20198838
Profile: Prof. Jae Kyoung Kim, PhD
jaekkim@kaist.ac.kr
http://mathsci.kaist.ac.kr/~jaekkim
Associate Professor
Department of Mathematical Sciences
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr Daejeon 34141, Korea
Profile: Dae Wook Kim, PhD Candidate
0308kdo@kaist.ac.kr
http://mathsci.kaist.ac.kr/~jaekkim
PhD Candidate
Department of Mathematical Sciences
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr Daejeon 34141, Korea
Profile: Dr. Cheng Chang, PhD
cheng.chang@pfizer.com
Associate Director of Clinical Pharmacology
Clinical Pharmacology, Global Product Development
Pfizer
https://www.pfizer.com/ Groton 06340, USA
(END)
2019.07.09 View 38222 -
 Deep Learning-Powered 'DeepEC' Helps Accurately Understand Enzyme Functions
(Figure: Overall scheme of DeepEC)
A deep learning-powered computational framework, ‘DeepEC,’ will allow the high-quality and high-throughput prediction of enzyme commission numbers, which is essential for the accurate understanding of enzyme functions.
A team of Dr. Jae Yong Ryu, Professor Hyun Uk Kim, and Distinguished Professor Sang Yup Lee at KAIST reported the computational framework powered by deep learning that predicts enzyme commission (EC) numbers with high precision in a high-throughput manner.
DeepEC takes a protein sequence as an input and accurately predicts EC numbers as an output. Enzymes are proteins that catalyze biochemical reactions and EC numbers consisting of four level numbers (i.e., a.b.c.d) indicate biochemical reactions. Thus, the identification of EC numbers is critical for accurately understanding enzyme functions and metabolism.
EC numbers are usually given to a protein sequence encoding an enzyme during a genome annotation procedure. Because of the importance of EC numbers, several EC number prediction tools have been developed, but they have room for further improvement with respect to computation time, precision, coverage, and the total size of the files needed for the EC number prediction.
DeepEC uses three convolutional neural networks (CNNs) as a major engine for the prediction of EC numbers, and also implements homology analysis for EC numbers if the three CNNs do not produce reliable EC numbers for a given protein sequence. DeepEC was developed by using a gold standard dataset covering 1,388,606 protein sequences and 4,669 EC numbers.
In particular, benchmarking studies of DeepEC and five other representative EC number prediction tools showed that DeepEC made the most precise and fastest predictions for EC numbers. DeepEC also required the smallest disk space for implementation, which makes it an ideal third-party software component.
Furthermore, DeepEC was the most sensitive in detecting enzymatic function loss as a result of mutations in domains/binding site residue of protein sequences; in this comparative analysis, all the domains or binding site residue were substituted with L-alanine residue in order to remove the protein function, which is known as the L-alanine scanning method.
This study was published online in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 20, 2019, entitled “Deep learning enables high-quality and high-throughput prediction of enzyme commission numbers.”
“DeepEC can be used as an independent tool and also as a third-party software component in combination with other computational platforms that examine metabolic reactions. DeepEC is freely available online,” said Professor Kim.
Distinguished Professor Lee said, “With DeepEC, it has become possible to process ever-increasing volumes of protein sequence data more efficiently and more accurately.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science and ICT through the National Research Foundation of Korea. This work was also funded by the Bio & Medical Technology Development Program of the National Research Foundation of Korea funded by the Korean government, the Ministry of Science and ICT.
Profile:
-Professor Hyun Uk Kim (ehukim@kaist.ac.kr)
https://sites.google.com/view/ehukim
Department of Chemical and Biomolecular Engineering
-Distinguished Professor Sang Yup Lee (leesy@kaist.ac.kr)
Department of Chemical and Biomolecular Engineering
http://mbel.kaist.ac.kr
2019.07.09 View 41112
Deep Learning-Powered 'DeepEC' Helps Accurately Understand Enzyme Functions
(Figure: Overall scheme of DeepEC)
A deep learning-powered computational framework, ‘DeepEC,’ will allow the high-quality and high-throughput prediction of enzyme commission numbers, which is essential for the accurate understanding of enzyme functions.
A team of Dr. Jae Yong Ryu, Professor Hyun Uk Kim, and Distinguished Professor Sang Yup Lee at KAIST reported the computational framework powered by deep learning that predicts enzyme commission (EC) numbers with high precision in a high-throughput manner.
DeepEC takes a protein sequence as an input and accurately predicts EC numbers as an output. Enzymes are proteins that catalyze biochemical reactions and EC numbers consisting of four level numbers (i.e., a.b.c.d) indicate biochemical reactions. Thus, the identification of EC numbers is critical for accurately understanding enzyme functions and metabolism.
EC numbers are usually given to a protein sequence encoding an enzyme during a genome annotation procedure. Because of the importance of EC numbers, several EC number prediction tools have been developed, but they have room for further improvement with respect to computation time, precision, coverage, and the total size of the files needed for the EC number prediction.
DeepEC uses three convolutional neural networks (CNNs) as a major engine for the prediction of EC numbers, and also implements homology analysis for EC numbers if the three CNNs do not produce reliable EC numbers for a given protein sequence. DeepEC was developed by using a gold standard dataset covering 1,388,606 protein sequences and 4,669 EC numbers.
In particular, benchmarking studies of DeepEC and five other representative EC number prediction tools showed that DeepEC made the most precise and fastest predictions for EC numbers. DeepEC also required the smallest disk space for implementation, which makes it an ideal third-party software component.
Furthermore, DeepEC was the most sensitive in detecting enzymatic function loss as a result of mutations in domains/binding site residue of protein sequences; in this comparative analysis, all the domains or binding site residue were substituted with L-alanine residue in order to remove the protein function, which is known as the L-alanine scanning method.
This study was published online in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 20, 2019, entitled “Deep learning enables high-quality and high-throughput prediction of enzyme commission numbers.”
“DeepEC can be used as an independent tool and also as a third-party software component in combination with other computational platforms that examine metabolic reactions. DeepEC is freely available online,” said Professor Kim.
Distinguished Professor Lee said, “With DeepEC, it has become possible to process ever-increasing volumes of protein sequence data more efficiently and more accurately.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science and ICT through the National Research Foundation of Korea. This work was also funded by the Bio & Medical Technology Development Program of the National Research Foundation of Korea funded by the Korean government, the Ministry of Science and ICT.
Profile:
-Professor Hyun Uk Kim (ehukim@kaist.ac.kr)
https://sites.google.com/view/ehukim
Department of Chemical and Biomolecular Engineering
-Distinguished Professor Sang Yup Lee (leesy@kaist.ac.kr)
Department of Chemical and Biomolecular Engineering
http://mbel.kaist.ac.kr
2019.07.09 View 41112 -
 Hydrogen-Natural Gas Hydrates Harvested by Natural Gas
A hydrogen-natural gas blend (HNGB) can be a game changer only if it can be stored safely and used as a sustainable clean energy resource. A recent study has suggested a new strategy for stably storing hydrogen, using natural gas as a stabilizer. The research proposed a practical gas phase modulator based synthesis of HNGB without generating chemical waste after dissociation for the immediate service.
The research team of Professor Jae Woo Lee from the Department of Chemical and Biomolecular Engineering in collaboration with the Gwangju Institute of Science and Technology (GIST) demonstrated that the natural gas modulator based synthesis leads to significantly reduced synthesis pressure simultaneously with the formation of hydrogen clusters in the confined nanoporous cages of clathrate hydrates. This approach minimizes the environmental impact and reduces operation costs since clathrate hydrates do not generate any chemical waste in both the synthesis and decomposition processes.
For the efficient storage and transportation of hydrogen, numerous materials have been investigated. Among others, clathrate hydrates offer distinct benefits. Clathrate hydrates are nanoporous inclusion compounds composed of a 3D network of polyhedral cages made of hydrogen-bonded ‘host’ water molecules and captured ‘guest’ gas or liquid molecules.
In this study, the research team used two gases, methane and ethane, which have lower equilibrium conditions compared to hydrogen as thermodynamic stabilizers. As a result, they succeeded in stably storing the hydrogen-natural gas compound in hydrates. According to the composition ratio of methane and ethane, structure I or II hydrates can be formed, both of which can stably store hydrogen-natural gas in low-pressure conditions.
The research team found that two hydrogen molecules are stored in small cages in tuned structure I hydrates, while up to three hydrogen molecules can be stored in both small and large cages in tuned structure II hydrates. Hydrates can store gas up to about 170-times its volume and the natural gas used as thermodynamic stabilizers in this study can also be used as an energy source.
The research team developed technology to produce hydrates from ice, produced hydrogen-natural gas hydrates by substitution, and successfully observed that the tuning phenomenon only occurs when hydrogen is involved in hydrate formation from the start for both structures of hydrates.
They expect that the findings can be applied to not only an energy-efficient gas storage material, but also a smart platform to utilize hydrogen natural gas blends, which can serve as a new alternative energy source with targeted hydrogen contents by designing synthetic pathways of mixed gas hydrates.
The research was published online in Energy Storage Materials on June 6, with the title ‘One-step formation of hydrogen clusters in clathrate hydrates stabilized via natural gas blending’.
Professor Lee said, “HNGB will utilize the existing natural gas infrastructure for transportation, so it is very likely that we can commercialize this hydrate system. We are investigating the kinetic performance through a follow-up strategy to increase the volume of gas storage.
This study was funded by the National Research Foundation of Korea and BK21 plus program.
(Figure1. Schematics showing the storage method for hydrogen in a natural gas hydrate using a substitution method and storage method directly from ice to a hydrogen-natural gas hydrate.)
(Figure 2. Artificially synthesized and dissociated hydrogen-natural gas hydrates. The Raman spectra of tuned sI and sII hydrate showing the hydrogen clusters in each cage.)
2019.06.21 View 43719
Hydrogen-Natural Gas Hydrates Harvested by Natural Gas
A hydrogen-natural gas blend (HNGB) can be a game changer only if it can be stored safely and used as a sustainable clean energy resource. A recent study has suggested a new strategy for stably storing hydrogen, using natural gas as a stabilizer. The research proposed a practical gas phase modulator based synthesis of HNGB without generating chemical waste after dissociation for the immediate service.
The research team of Professor Jae Woo Lee from the Department of Chemical and Biomolecular Engineering in collaboration with the Gwangju Institute of Science and Technology (GIST) demonstrated that the natural gas modulator based synthesis leads to significantly reduced synthesis pressure simultaneously with the formation of hydrogen clusters in the confined nanoporous cages of clathrate hydrates. This approach minimizes the environmental impact and reduces operation costs since clathrate hydrates do not generate any chemical waste in both the synthesis and decomposition processes.
For the efficient storage and transportation of hydrogen, numerous materials have been investigated. Among others, clathrate hydrates offer distinct benefits. Clathrate hydrates are nanoporous inclusion compounds composed of a 3D network of polyhedral cages made of hydrogen-bonded ‘host’ water molecules and captured ‘guest’ gas or liquid molecules.
In this study, the research team used two gases, methane and ethane, which have lower equilibrium conditions compared to hydrogen as thermodynamic stabilizers. As a result, they succeeded in stably storing the hydrogen-natural gas compound in hydrates. According to the composition ratio of methane and ethane, structure I or II hydrates can be formed, both of which can stably store hydrogen-natural gas in low-pressure conditions.
The research team found that two hydrogen molecules are stored in small cages in tuned structure I hydrates, while up to three hydrogen molecules can be stored in both small and large cages in tuned structure II hydrates. Hydrates can store gas up to about 170-times its volume and the natural gas used as thermodynamic stabilizers in this study can also be used as an energy source.
The research team developed technology to produce hydrates from ice, produced hydrogen-natural gas hydrates by substitution, and successfully observed that the tuning phenomenon only occurs when hydrogen is involved in hydrate formation from the start for both structures of hydrates.
They expect that the findings can be applied to not only an energy-efficient gas storage material, but also a smart platform to utilize hydrogen natural gas blends, which can serve as a new alternative energy source with targeted hydrogen contents by designing synthetic pathways of mixed gas hydrates.
The research was published online in Energy Storage Materials on June 6, with the title ‘One-step formation of hydrogen clusters in clathrate hydrates stabilized via natural gas blending’.
Professor Lee said, “HNGB will utilize the existing natural gas infrastructure for transportation, so it is very likely that we can commercialize this hydrate system. We are investigating the kinetic performance through a follow-up strategy to increase the volume of gas storage.
This study was funded by the National Research Foundation of Korea and BK21 plus program.
(Figure1. Schematics showing the storage method for hydrogen in a natural gas hydrate using a substitution method and storage method directly from ice to a hydrogen-natural gas hydrate.)
(Figure 2. Artificially synthesized and dissociated hydrogen-natural gas hydrates. The Raman spectra of tuned sI and sII hydrate showing the hydrogen clusters in each cage.)
2019.06.21 View 43719