Department+of+Physics
-
 A New Approach to 3D Holographic Displays Greatly Improves the Image Quality
With the addition of holographic diffusers or frosted glasses to wavefront modulators, KAIST researchers offer a simple and practical solution to significantly enhance the performance of 3D dynamic holographic displays by 2,600 times.
The potential applications of three-dimensional (3D) digital holograms are enormous. In addition to arts and entertainment, various fields including biomedical imaging, scientific visualization, engineering design, and displays could benefit from this technology. For example, creating full-sized organs for 3D analysis by doctors could be helpful, but it remained a challenge owing to the limitation of hologram-generation techniques.
A research team led by Professor YongKeun Park of the Physics Department at KAIST has come up with a solution and developed a 3D holographic display that performs more than 2,600 times better than existing 3D holographic displays. This study is expected to improve the limited size and viewing angle of 3D images, which were a major problem of the current holographic displays. The study was published online in Nature Photonics on January 23, 2017.
3D holograms, which often appear in science fiction films, are a familiar technology to the public, but holograms in movies are created with computer graphic effects. Methods for creating true 3D holograms are still being studied in the laboratory. For example, due to the difficulty of generating real 3D images, recent virtual reality (VR) and augmented reality (AR) devices project two different two-dimensional (2D) images onto a viewer to induce optical illusions.
To create a 3D hologram that can be viewed without special equipment such as 3D glasses, the wavefront of light must be controlled using wavefront modulators such as spatial light modulators (SLMs) and deformable mirrors (DMs). A wavefront modulator is an optical manipulation device that can control the direction of light propagation.
However, the biggest limitation to using these modulators as 3D displays is the number of pixels. The large number of pixels that are packed into high-resolution displays developed in recent years are suitable for a 2D image, and the amount of information contained in those pixels cannot produce a 3D image. For this reason, a 3D image that can be made with existing wavefront modulator technology is 1 cm in size with a narrow viewing angle of 3 degrees, which is far from practicable.
As an alternative, KAIST researchers used a DM and added two successive holographic diffusers to scatter light. By scattering light in many directions, this allows for a wider viewing angle and larger image, but results in volume speckle fields, which are caused by the interference of multiple scattered light. Random volume speckle fields cannot be used to display 3D images.
To fix the problem, the researchers employed a wavefront-shaping technique to control the fields. As a result, they succeeded in producing an enhanced 3D holographic image with a viewing angle of 35 degrees in a volume of 2 cm in length, width, and height. This yielded a performance that was about 2,600 times stronger than the original image definition generated when they used a DM without a diffuser.
Professor Park said, “Scattering light has previously been believed to interfere with the recognition of objects, but we have demonstrated that current 3D displays can be improved significantly with an increased viewing angle and image size by properly controlling the scattered light.”
Hyeonseung Yu, who is the lead author of this research article and a doctoral candidate in the Department of Physics, KAIST, noted that this technology signals a good start to develop a practical model for dynamic 3D hologram displays that can be enjoyed without the need for special eyeglasses. “This approach can also be applied to AR and VR technology to enhance the image resolution and viewing angles,” added Yu.
The research paper is entitled “Ultrahigh-definition Dynamic 3D Holographic Display by Active Control of Volume Speckle Fields.”
Figure 1. Concept of Scattering Display
The size and viewing angle of 3D images can be simultaneously increased when a scattering medium (diffuser) is introduced. By controlling the wavefront impinging on the scattering medium, the desired 3D hologram is generated.
Figure 2. Experimental Setup
The optical set-up consists of a deformable mirror and the scattering medium with two successive holographic diffusers. A high-numerical-aperture imaging unit mounted on a three-axis motorized translational system is utilized for wavefront optimization and imaging.
Figure 3. 3D Images Projected
This picture shows 3D images in a volume of 2 cm × 2 cm × 2 cm with a viewing angle of 35 degrees using one of the wavefront modulators, a digital micromirror device (DMD).
Figure 4. Artist’s Rendition of the Proposed Concept
A dynamic 3D hologram of a face is displayed.
2017.02.01 View 14841
A New Approach to 3D Holographic Displays Greatly Improves the Image Quality
With the addition of holographic diffusers or frosted glasses to wavefront modulators, KAIST researchers offer a simple and practical solution to significantly enhance the performance of 3D dynamic holographic displays by 2,600 times.
The potential applications of three-dimensional (3D) digital holograms are enormous. In addition to arts and entertainment, various fields including biomedical imaging, scientific visualization, engineering design, and displays could benefit from this technology. For example, creating full-sized organs for 3D analysis by doctors could be helpful, but it remained a challenge owing to the limitation of hologram-generation techniques.
A research team led by Professor YongKeun Park of the Physics Department at KAIST has come up with a solution and developed a 3D holographic display that performs more than 2,600 times better than existing 3D holographic displays. This study is expected to improve the limited size and viewing angle of 3D images, which were a major problem of the current holographic displays. The study was published online in Nature Photonics on January 23, 2017.
3D holograms, which often appear in science fiction films, are a familiar technology to the public, but holograms in movies are created with computer graphic effects. Methods for creating true 3D holograms are still being studied in the laboratory. For example, due to the difficulty of generating real 3D images, recent virtual reality (VR) and augmented reality (AR) devices project two different two-dimensional (2D) images onto a viewer to induce optical illusions.
To create a 3D hologram that can be viewed without special equipment such as 3D glasses, the wavefront of light must be controlled using wavefront modulators such as spatial light modulators (SLMs) and deformable mirrors (DMs). A wavefront modulator is an optical manipulation device that can control the direction of light propagation.
However, the biggest limitation to using these modulators as 3D displays is the number of pixels. The large number of pixels that are packed into high-resolution displays developed in recent years are suitable for a 2D image, and the amount of information contained in those pixels cannot produce a 3D image. For this reason, a 3D image that can be made with existing wavefront modulator technology is 1 cm in size with a narrow viewing angle of 3 degrees, which is far from practicable.
As an alternative, KAIST researchers used a DM and added two successive holographic diffusers to scatter light. By scattering light in many directions, this allows for a wider viewing angle and larger image, but results in volume speckle fields, which are caused by the interference of multiple scattered light. Random volume speckle fields cannot be used to display 3D images.
To fix the problem, the researchers employed a wavefront-shaping technique to control the fields. As a result, they succeeded in producing an enhanced 3D holographic image with a viewing angle of 35 degrees in a volume of 2 cm in length, width, and height. This yielded a performance that was about 2,600 times stronger than the original image definition generated when they used a DM without a diffuser.
Professor Park said, “Scattering light has previously been believed to interfere with the recognition of objects, but we have demonstrated that current 3D displays can be improved significantly with an increased viewing angle and image size by properly controlling the scattered light.”
Hyeonseung Yu, who is the lead author of this research article and a doctoral candidate in the Department of Physics, KAIST, noted that this technology signals a good start to develop a practical model for dynamic 3D hologram displays that can be enjoyed without the need for special eyeglasses. “This approach can also be applied to AR and VR technology to enhance the image resolution and viewing angles,” added Yu.
The research paper is entitled “Ultrahigh-definition Dynamic 3D Holographic Display by Active Control of Volume Speckle Fields.”
Figure 1. Concept of Scattering Display
The size and viewing angle of 3D images can be simultaneously increased when a scattering medium (diffuser) is introduced. By controlling the wavefront impinging on the scattering medium, the desired 3D hologram is generated.
Figure 2. Experimental Setup
The optical set-up consists of a deformable mirror and the scattering medium with two successive holographic diffusers. A high-numerical-aperture imaging unit mounted on a three-axis motorized translational system is utilized for wavefront optimization and imaging.
Figure 3. 3D Images Projected
This picture shows 3D images in a volume of 2 cm × 2 cm × 2 cm with a viewing angle of 35 degrees using one of the wavefront modulators, a digital micromirror device (DMD).
Figure 4. Artist’s Rendition of the Proposed Concept
A dynamic 3D hologram of a face is displayed.
2017.02.01 View 14841 -
 Technology to Allow Non-Magnetic Materials to Have Magnetic Properties by Professor Chan-Ho Yang
Professor Chan-Ho Yang and his research team from the Department of Physics at KAIST have developed a technology that allows non-magnetic materials to have magnetic properties or, in reverse, to remove magnetic properties from a magnet using an electric field.
Based on this research, it is expected that if magnetic-material-based data storage is developed, applications for high-speed massive data transfer will be possible.
The results of this research, with Ph.D. candidate Byung-Kwon Jang as the first author, were published online in Nature Physics on October 3.
Very small magnets exist inside of any materials. If the direction of the minuscule magnets is dis-aligned, pointing multiple directions, it is non-magnetic. If the direction is aligned in a certain direction, the material holds magnetic property just like any magnet we normally see.
Data storage capacity technology has rapidly advanced to the point where we can easily get a portable hard disk drive (HDD) with terabyte-level storage; however, the increase in storage is inevitably followed by slower data access speed for a storage device. Although HDDs are currently the most widely used data storage devices, their technical applications are limited due to their slow data access speed.
Other methods such as solid-state drives (SSDs), floating gates, and resistive switching have been developed as alternatives. Yet, they leave tracks every time data is written, and this can cause fatigue cumulative damage.
There have been many attempts to compose cells—the smallest data storage space on a storage device—with magnetic materials as that would enable faster data access speeds and remove fatigue cumulative damage. Generally, the techniques tried by researchers were to use induced magnetic fields through current flow. However, magnetic fields are very difficult to shield and can affect a large area. As a result, they alternate the magnetic property of adjacent cells. Because each cell cannot be adjusted one by one, it cannot also be arranged in a certain direction, and therefore, it is hard to change the magnetic state.
Professor Yang and his team adjusted the magnetic state by using magnetoelectric interaction to deal with this issue. Instead of using magnetic fields, magnetoelectric interaction is a method that uses an electric field to adjust the magnetic state. It has the advantage of smaller energy consumption as well.
Professor Yang's team demonstrated that cells facing random directions can be arranged in a certain direction by only inducing an electric field. In addition, the reverse was also proved to be feasible.
Until this research, most cases of previous findings were only feasible at extremely low temperatures or high temperatures, but the technology developed by the research team is practicable at room temperature by manipulating chemical pressure. It allows for a reversible magnetic state, and moreover, is non-volatile. Therefore, the results of this research are expected to provide the basis for developing next-generation information storage device.
Professor Yang said, “The changes in the electric magnetic state will be accompanied by entropy changes” and added, “Our research is expected to open new potential for future applications not only for magnetoelectric devices, but also for thermoelectric effect.”
This research has been worked on jointly with Dr. Si-Yong Choi from the Korea Institute of Materials Science, Prof. Yoon-Hee Jeong from the Pohang University of Science and Technology, Dr. Tae-Yeong Koo from the Pohang Accelerator Laboratory, Dr. Kyung-Tae Ko from the Max Planck Institute for Chemical Physics of Solids, Dr. Jun-Sik Lee and Dr. Hendrik Ohldag from the SLAC National Accelerator Laboratory of the United States, and Prof. Jan Seidel from the University of New South Wales of Australia.
The research was supported by the Mid-Career Researcher Program of the National Research Foundation of Korea, Global Research Network Support Project, Leading Research Center Support Project (Condensed Quantum Coherence Research Center), Global Frontier Project (Hybrid Interface Materials Research Group), and others.
Picture: The concept graphic for the electric-field-induced magnetic phase switching the magnetic direction
2016.11.04 View 8944
Technology to Allow Non-Magnetic Materials to Have Magnetic Properties by Professor Chan-Ho Yang
Professor Chan-Ho Yang and his research team from the Department of Physics at KAIST have developed a technology that allows non-magnetic materials to have magnetic properties or, in reverse, to remove magnetic properties from a magnet using an electric field.
Based on this research, it is expected that if magnetic-material-based data storage is developed, applications for high-speed massive data transfer will be possible.
The results of this research, with Ph.D. candidate Byung-Kwon Jang as the first author, were published online in Nature Physics on October 3.
Very small magnets exist inside of any materials. If the direction of the minuscule magnets is dis-aligned, pointing multiple directions, it is non-magnetic. If the direction is aligned in a certain direction, the material holds magnetic property just like any magnet we normally see.
Data storage capacity technology has rapidly advanced to the point where we can easily get a portable hard disk drive (HDD) with terabyte-level storage; however, the increase in storage is inevitably followed by slower data access speed for a storage device. Although HDDs are currently the most widely used data storage devices, their technical applications are limited due to their slow data access speed.
Other methods such as solid-state drives (SSDs), floating gates, and resistive switching have been developed as alternatives. Yet, they leave tracks every time data is written, and this can cause fatigue cumulative damage.
There have been many attempts to compose cells—the smallest data storage space on a storage device—with magnetic materials as that would enable faster data access speeds and remove fatigue cumulative damage. Generally, the techniques tried by researchers were to use induced magnetic fields through current flow. However, magnetic fields are very difficult to shield and can affect a large area. As a result, they alternate the magnetic property of adjacent cells. Because each cell cannot be adjusted one by one, it cannot also be arranged in a certain direction, and therefore, it is hard to change the magnetic state.
Professor Yang and his team adjusted the magnetic state by using magnetoelectric interaction to deal with this issue. Instead of using magnetic fields, magnetoelectric interaction is a method that uses an electric field to adjust the magnetic state. It has the advantage of smaller energy consumption as well.
Professor Yang's team demonstrated that cells facing random directions can be arranged in a certain direction by only inducing an electric field. In addition, the reverse was also proved to be feasible.
Until this research, most cases of previous findings were only feasible at extremely low temperatures or high temperatures, but the technology developed by the research team is practicable at room temperature by manipulating chemical pressure. It allows for a reversible magnetic state, and moreover, is non-volatile. Therefore, the results of this research are expected to provide the basis for developing next-generation information storage device.
Professor Yang said, “The changes in the electric magnetic state will be accompanied by entropy changes” and added, “Our research is expected to open new potential for future applications not only for magnetoelectric devices, but also for thermoelectric effect.”
This research has been worked on jointly with Dr. Si-Yong Choi from the Korea Institute of Materials Science, Prof. Yoon-Hee Jeong from the Pohang University of Science and Technology, Dr. Tae-Yeong Koo from the Pohang Accelerator Laboratory, Dr. Kyung-Tae Ko from the Max Planck Institute for Chemical Physics of Solids, Dr. Jun-Sik Lee and Dr. Hendrik Ohldag from the SLAC National Accelerator Laboratory of the United States, and Prof. Jan Seidel from the University of New South Wales of Australia.
The research was supported by the Mid-Career Researcher Program of the National Research Foundation of Korea, Global Research Network Support Project, Leading Research Center Support Project (Condensed Quantum Coherence Research Center), Global Frontier Project (Hybrid Interface Materials Research Group), and others.
Picture: The concept graphic for the electric-field-induced magnetic phase switching the magnetic direction
2016.11.04 View 8944 -
 Next-Generation Holographic Microscope for 3D Live Cell Imaging
KAIST researchers have developed a revolutionary bio-medical imaging tool, the HT-1, to view and analyze cells, which is commercially available.
Professor YongKeun Park of the Physics Department at KAIST and his research team have developed a powerful method for 3D imaging of live cells without staining. The researchers announced the launch of their new microscopic tool, the holotomography (HT)-1, to the global marketplace through a Korean start-up that Professor Park co-founded, TomoCube (www.tomocube.com).
Professor Park is a leading researcher in the field of biophotonics and has dedicated much of his research career to working on digital holographic microscopy technology. He collaborated with TomoCube’s R&D team to develop a state-of-the-art, 2D/3D/4D holographic microscope that would allow a real-time label-free visualization of biological cells and tissues.
The HT is an optical analogy of X-ray computed tomography (CT). Both X-ray CT and HT share the same physical principle—the inverse of wave scattering. The difference is that HT uses laser illumination whereas X-ray CT uses X-ray beams. From the measurement of multiple 2D holograms of a cell, coupled with various angles of laser illuminations, the 3D refractive index (RI) distribution of the cell can be reconstructed. The reconstructed 3D RI map provides structural and chemical information of the cell including mass, morphology, protein concentration, and dynamics of the cellular membrane.
The HT enables users to quantitatively and non-invasively investigate the intrinsic properties of biological cells, for example, dry mass and protein concentration. Some of the research team’s breakthroughs that have leveraged HT’s unique and special capabilities can be found in several recent publications, including a lead article on the simultaneous 3D visualization and position tracking of optically trapped particles which was published in Optica on April 20, 2015.
Current fluorescence confocal microscopy techniques require the use of exogenous labeling agents to render high-contrast molecular information. Therefore, drawbacks include possible photo-bleaching, photo-toxicity, and interference with normal molecular activities. Immune or stem cells that need to be reinjected into the body are considered particularly difficult to employ with fluorescence microscopy.
“As one of the two currently available, high-resolution tomographic microscopes in the world, I believe that the HT-1 is the best in class regarding specifications and functionality. Users can see 3D/4D live images of cells, without fixing, coating or staining cells. Sample preparation times are reduced from a few days or hours to just a few minutes,” said Professor Park.
Two Korean hospitals, Seoul National University Hospital in Bundang and Boramae Hospital in Seoul, are using this microscope currently. The research team has also introduced the HT-1 at the Photonics West Exhibition 2016 that took place on February 16-18 in San Francisco, USA.
Professor Park added, “Our technology has set a new paradigm for cell observation under a microscope. I expect that this tomographic microscopy will be more widely used in future in various areas of pharmaceuticals, neuroscience, immunology, hematology, and cell biology.”
Figure 1: HT-1 and Its Specifications
Figure 2: 3D Images of Representative Biological Cells Taken with the HT-1
2016.03.29 View 14590
Next-Generation Holographic Microscope for 3D Live Cell Imaging
KAIST researchers have developed a revolutionary bio-medical imaging tool, the HT-1, to view and analyze cells, which is commercially available.
Professor YongKeun Park of the Physics Department at KAIST and his research team have developed a powerful method for 3D imaging of live cells without staining. The researchers announced the launch of their new microscopic tool, the holotomography (HT)-1, to the global marketplace through a Korean start-up that Professor Park co-founded, TomoCube (www.tomocube.com).
Professor Park is a leading researcher in the field of biophotonics and has dedicated much of his research career to working on digital holographic microscopy technology. He collaborated with TomoCube’s R&D team to develop a state-of-the-art, 2D/3D/4D holographic microscope that would allow a real-time label-free visualization of biological cells and tissues.
The HT is an optical analogy of X-ray computed tomography (CT). Both X-ray CT and HT share the same physical principle—the inverse of wave scattering. The difference is that HT uses laser illumination whereas X-ray CT uses X-ray beams. From the measurement of multiple 2D holograms of a cell, coupled with various angles of laser illuminations, the 3D refractive index (RI) distribution of the cell can be reconstructed. The reconstructed 3D RI map provides structural and chemical information of the cell including mass, morphology, protein concentration, and dynamics of the cellular membrane.
The HT enables users to quantitatively and non-invasively investigate the intrinsic properties of biological cells, for example, dry mass and protein concentration. Some of the research team’s breakthroughs that have leveraged HT’s unique and special capabilities can be found in several recent publications, including a lead article on the simultaneous 3D visualization and position tracking of optically trapped particles which was published in Optica on April 20, 2015.
Current fluorescence confocal microscopy techniques require the use of exogenous labeling agents to render high-contrast molecular information. Therefore, drawbacks include possible photo-bleaching, photo-toxicity, and interference with normal molecular activities. Immune or stem cells that need to be reinjected into the body are considered particularly difficult to employ with fluorescence microscopy.
“As one of the two currently available, high-resolution tomographic microscopes in the world, I believe that the HT-1 is the best in class regarding specifications and functionality. Users can see 3D/4D live images of cells, without fixing, coating or staining cells. Sample preparation times are reduced from a few days or hours to just a few minutes,” said Professor Park.
Two Korean hospitals, Seoul National University Hospital in Bundang and Boramae Hospital in Seoul, are using this microscope currently. The research team has also introduced the HT-1 at the Photonics West Exhibition 2016 that took place on February 16-18 in San Francisco, USA.
Professor Park added, “Our technology has set a new paradigm for cell observation under a microscope. I expect that this tomographic microscopy will be more widely used in future in various areas of pharmaceuticals, neuroscience, immunology, hematology, and cell biology.”
Figure 1: HT-1 and Its Specifications
Figure 2: 3D Images of Representative Biological Cells Taken with the HT-1
2016.03.29 View 14590 -
 Mapping the Folding Process of a Single Membrane Protein
KAIST and UCLA scientists were able to observe an individual membrane protein fold and unfold by pulling and releasing magnetically trapped protein molecules.
Proteins are huge molecules containing hundreds to thousands of atoms that adopt a unique three dimensional structure, placing chemical groups in just the right place to catalyze reactions or build cellular structures. How all those atoms manage to find the right location - the so-called folding problem - has fascinated molecular biologists since the first structures were seen in the 1950s. Moreover, folding has important medical implications because most genetic defects cause protein misfolding.
About a third of all proteins float around in the cell membrane where they ensure the right chemicals get in the cell in the right amounts. Membrane proteins also provide key information links between the cell and its environment. Indeed, most drugs target membrane proteins. Nevertheless, the folding of membrane proteins has been particularly difficult to study and has rarely been studied in natural environments, leaving the folding process for a large fraction of the protein universe still largely cloaked in mystery.
In a recent issue of Nature Chemical Biology, published on October 19, 2015, a research team led by Tae-Young Yoon of the Department of Physics at the Korea Advanced Institute of Science and Technology (KAIST) and James U. Bowie of the Department of Chemistry and Biochemistry at the University of California, Los Angeles (UCLA), report a new method for manipulating the folding of membrane proteins in a membrane environment using a tool called a magnetic tweezer.
Researchers first attach long DNA handles to the ends of the protein. One handle is attached to a glass surface and the other to a magnetic bead. Using a magnet, they can essentially grab the protein and pull on it, inducing it to unfold. By playing with the bead attached to the protein, they can force the protein to unfold or allow it to refold, and watch all this happening by 3D-tracking of the magnetic bead. With this novel strategy, they were able to quantitatively map the folding energy landscape, the folding kinetic rate, and folding intermediates of a membrane protein in a membrane environment for the first time.
“I have been dreaming about this experiment for a decade. To see it work so well is really gratifying,” said Dr. Bowie.
One of the major surprises in the study was that essentially all the atoms of the protein jump into the correct structure together. The researchers expected that the protein structure would come together in a more piecemeal fashion, with different parts of the structure forming separately, but that was not the case. It is possible that nature evolved such a smooth, highly cooperative folding process to prevent partially folded forms that could get into trouble in the crowded cell membrane. On the other hand, the cooperative folding seen here might not apply to other membrane proteins.
“We need to look at more proteins. The technique developed here may allow us to do just that,” said Dr. Yoon.
The single molecule mechanical manipulation technique could enable detailed folding studies of many other membrane proteins. A major barrier to the study of membrane proteins previously is that the proteins tend to stick together and get tangled up, as computer cords lying at your feet tend to do. With the tweezer technique used in this work, the protein cords are held apart from other cords so they can’t get knotted up. It is hoped that the new approach will open up an important part of the protein universe to scrutiny, including many proteins that become misfolded in disease states.
The title of the research paper is “Mapping the energy landscape for second-stage folding of a single membrane protein” (DOI: 10.1038/nchembio.1939).
Picture: Single-molecule magnetic tweezers that induce mechanical unfolding and refolding of a single membrane protein. Since the force applied is parallel to the biological lipid membrane, the unfolding and refolding processes occur within the membrane.
2015.10.20 View 11030
Mapping the Folding Process of a Single Membrane Protein
KAIST and UCLA scientists were able to observe an individual membrane protein fold and unfold by pulling and releasing magnetically trapped protein molecules.
Proteins are huge molecules containing hundreds to thousands of atoms that adopt a unique three dimensional structure, placing chemical groups in just the right place to catalyze reactions or build cellular structures. How all those atoms manage to find the right location - the so-called folding problem - has fascinated molecular biologists since the first structures were seen in the 1950s. Moreover, folding has important medical implications because most genetic defects cause protein misfolding.
About a third of all proteins float around in the cell membrane where they ensure the right chemicals get in the cell in the right amounts. Membrane proteins also provide key information links between the cell and its environment. Indeed, most drugs target membrane proteins. Nevertheless, the folding of membrane proteins has been particularly difficult to study and has rarely been studied in natural environments, leaving the folding process for a large fraction of the protein universe still largely cloaked in mystery.
In a recent issue of Nature Chemical Biology, published on October 19, 2015, a research team led by Tae-Young Yoon of the Department of Physics at the Korea Advanced Institute of Science and Technology (KAIST) and James U. Bowie of the Department of Chemistry and Biochemistry at the University of California, Los Angeles (UCLA), report a new method for manipulating the folding of membrane proteins in a membrane environment using a tool called a magnetic tweezer.
Researchers first attach long DNA handles to the ends of the protein. One handle is attached to a glass surface and the other to a magnetic bead. Using a magnet, they can essentially grab the protein and pull on it, inducing it to unfold. By playing with the bead attached to the protein, they can force the protein to unfold or allow it to refold, and watch all this happening by 3D-tracking of the magnetic bead. With this novel strategy, they were able to quantitatively map the folding energy landscape, the folding kinetic rate, and folding intermediates of a membrane protein in a membrane environment for the first time.
“I have been dreaming about this experiment for a decade. To see it work so well is really gratifying,” said Dr. Bowie.
One of the major surprises in the study was that essentially all the atoms of the protein jump into the correct structure together. The researchers expected that the protein structure would come together in a more piecemeal fashion, with different parts of the structure forming separately, but that was not the case. It is possible that nature evolved such a smooth, highly cooperative folding process to prevent partially folded forms that could get into trouble in the crowded cell membrane. On the other hand, the cooperative folding seen here might not apply to other membrane proteins.
“We need to look at more proteins. The technique developed here may allow us to do just that,” said Dr. Yoon.
The single molecule mechanical manipulation technique could enable detailed folding studies of many other membrane proteins. A major barrier to the study of membrane proteins previously is that the proteins tend to stick together and get tangled up, as computer cords lying at your feet tend to do. With the tweezer technique used in this work, the protein cords are held apart from other cords so they can’t get knotted up. It is hoped that the new approach will open up an important part of the protein universe to scrutiny, including many proteins that become misfolded in disease states.
The title of the research paper is “Mapping the energy landscape for second-stage folding of a single membrane protein” (DOI: 10.1038/nchembio.1939).
Picture: Single-molecule magnetic tweezers that induce mechanical unfolding and refolding of a single membrane protein. Since the force applied is parallel to the biological lipid membrane, the unfolding and refolding processes occur within the membrane.
2015.10.20 View 11030 -
 3D Plasmon Antenna Capable of Focusing Light into Few Nanometers
Professors Myung-Ki Kim and Yong-Hee Lee, both of the Physics Department at KAIST, and their research teams have developed a three dimensional (3D) gap-plasmon antenna which can focus light into a space a few nanometers wide. Their research findings were published in the June 10th issue of Nano Letters.
Focusing light into a point-like space is an active research field with many applications. However, concentrating light into a smaller space than its wavelength is often hindered by diffraction. To tackle this problem, many researchers have utilized the plasmonic phenomenon of a metal where light can be confined to a greater extent by overcoming the diffraction limit.
Many researchers have focused on developing a two dimensional (2D) plasmon antenna and were able to focus a light under 5 nanometers wide. However, this 2D antenna revealed a challenge: the light disperses to the opposite end regardless of how small its beam was focused. To solve this difficulty, a 3D structure had to be employed to maximize the light's intensity.
Adopting the proximal focused-ion-beam milling technology, the KAIST research team developed a 3D four nanometer wide gap-plasmon antenna. By squeezing the photons into a 3D nano space of 4 x 10 x 10 nm3 size, the researchers were able to increase the intensity of light by 400,000 times stronger than that of the incident light. Capitalizing on the enhanced intensity of light within the antenna, they intensified the second-harmonic signal and verified that the light was focused in the nano gap by scanning cathodoluminescent images.
The researchers anticipate that this technology will improve the speed of data transfer and processing up to the level of a terahertz (one trillion times per second) and to enlarge the storage volume per unit area on hard disks by 100 times. In addition, high definition images of submolecule size can be taken with actual light, instead of with an electron microscope, while improving the semiconductor process to a smaller size of few nanometers.
Professor Kim said, “A simple yet ingenious idea has shifted the research paradigm from 2D gap-plasmon antennas to 3D antennas. This technology will see numerous applications including in the field of information technology, data storage, imaging medical science, and semiconductor processes.”
The research was sponsored by the National Research Foundation of Korea.
Figure 1: 3D Gap-Plasmon Antenna Structure and Simulation Results
Figure 2 – Constructed 3D Gap-Plasmon Antenna Structure
Figure 3 – Amplified Second Harmonic Signal Generation and Light Focused in the Nano Gap
2015.06.24 View 11616
3D Plasmon Antenna Capable of Focusing Light into Few Nanometers
Professors Myung-Ki Kim and Yong-Hee Lee, both of the Physics Department at KAIST, and their research teams have developed a three dimensional (3D) gap-plasmon antenna which can focus light into a space a few nanometers wide. Their research findings were published in the June 10th issue of Nano Letters.
Focusing light into a point-like space is an active research field with many applications. However, concentrating light into a smaller space than its wavelength is often hindered by diffraction. To tackle this problem, many researchers have utilized the plasmonic phenomenon of a metal where light can be confined to a greater extent by overcoming the diffraction limit.
Many researchers have focused on developing a two dimensional (2D) plasmon antenna and were able to focus a light under 5 nanometers wide. However, this 2D antenna revealed a challenge: the light disperses to the opposite end regardless of how small its beam was focused. To solve this difficulty, a 3D structure had to be employed to maximize the light's intensity.
Adopting the proximal focused-ion-beam milling technology, the KAIST research team developed a 3D four nanometer wide gap-plasmon antenna. By squeezing the photons into a 3D nano space of 4 x 10 x 10 nm3 size, the researchers were able to increase the intensity of light by 400,000 times stronger than that of the incident light. Capitalizing on the enhanced intensity of light within the antenna, they intensified the second-harmonic signal and verified that the light was focused in the nano gap by scanning cathodoluminescent images.
The researchers anticipate that this technology will improve the speed of data transfer and processing up to the level of a terahertz (one trillion times per second) and to enlarge the storage volume per unit area on hard disks by 100 times. In addition, high definition images of submolecule size can be taken with actual light, instead of with an electron microscope, while improving the semiconductor process to a smaller size of few nanometers.
Professor Kim said, “A simple yet ingenious idea has shifted the research paradigm from 2D gap-plasmon antennas to 3D antennas. This technology will see numerous applications including in the field of information technology, data storage, imaging medical science, and semiconductor processes.”
The research was sponsored by the National Research Foundation of Korea.
Figure 1: 3D Gap-Plasmon Antenna Structure and Simulation Results
Figure 2 – Constructed 3D Gap-Plasmon Antenna Structure
Figure 3 – Amplified Second Harmonic Signal Generation and Light Focused in the Nano Gap
2015.06.24 View 11616 -
 Fast, Accurate 3D Imaging to Track Optically-Trapped Particles
KAIST researchers published an article on the development of a novel technique to precisely track the 3-D positions of optically-trapped particles having complicated geometry in high speed in the April 2015 issue of Optica.
Optical tweezers have been used as an invaluable tool for exerting micro-scale force on microscopic particles and manipulating three-dimensional (3-D) positions of particles. Optical tweezers employ a tightly-focused laser whose beam diameter is smaller than one micrometer (1/100 of hair thickness), which generates attractive force on neighboring microscopic particles moving toward the beam focus. Controlling the positions of the beam focus enabled researchers to hold the particles and move them freely to other locations so they coined the name “optical tweezers.”
To locate the optically-trapped particles by a laser beam, optical microscopes have usually been employed. Optical microscopes measure light signals scattered by the optically-trapped microscopic particles and the positions of the particles in two dimensions. However, it was difficult to quantify the particles’ precise positions along the optic axis, the direction of the beam, from a single image, which is analogous to the difficulty of determining the front and rear positions of objects when closing an eye due to a lack of depth perception. Furthermore, it became more difficult to measure precisely 3-D positions of particles when scattered light signals were distorted by optically-trapped particles having complicated shapes or other particles occlude the target object along the optic axis.
Professor YongKeun Park and his research team in the Department of Physics at the Korea Advanced Institute of Science and Technology (KAIST) employed an optical diffraction tomography (ODT) technique to measure 3-D positions of optically-trapped particles in high speed. The principle of ODT is similar to X-ray CT imaging commonly used in hospitals for visualizing the internal organs of patients. Like X-ray CT imaging, which takes several images from various illumination angles, ODT measures 3-D images of optically-trapped particles by illuminating them with a laser beam in various incidence angles.
The KAIST team used optical tweezers to trap a glass bead with a diameter of 2 micrometers, and moved the bead toward a white blood cell having complicated internal structures. The team measured the 3-D dynamics of the white blood cell as it responded to an approaching glass bead via ODT in the high acquisition rate of 60 images per second. Since the white blood cell screens the glass bead along an optic axis, a conventionally-used optical microscope could not determine the 3-D positions of the glass bead. In contrast, the present method employing ODT localized the 3-D positions of the bead precisely as well as measured the composition of the internal materials of the bead and the white blood cell simultaneously.
Professor Park said, “Our technique has the advantage of measuring the 3-D positions and internal structures of optically-trapped particles in high speed without labelling exogenous fluorescent agents and can be applied in various fields including physics, optics, nanotechnology, and medical science.”
Kyoohyun Kim, the lead author of this paper (“Simultaneous 3D Visualization and Position Tracking of Optically Trapped Particles Using Optical Diffraction Tomography”), added, “This ODT technique can also apply to cellular-level surgeries where optical tweezers are used to manipulate intracellular organelles and to display in real time and in 3-D the images of the reaction of the cell membrane and nucleus during the operation or monitoring the recovery process of the cells from the surgery.”
The research results were published as the cover article in the April 2014 issue of Optica, the newest journal launched last year by the Optical Society of America (OSA) for rapid dissemination of high-impact results related to optics.
Figure 1: This picture shows the concept image of tweezing an optically-trapped glass bead on the cellular membrane of a white blood cell.
Figure 2: High-speed 3-D images produced from optical diffraction tomography technique
2015.04.24 View 13173
Fast, Accurate 3D Imaging to Track Optically-Trapped Particles
KAIST researchers published an article on the development of a novel technique to precisely track the 3-D positions of optically-trapped particles having complicated geometry in high speed in the April 2015 issue of Optica.
Optical tweezers have been used as an invaluable tool for exerting micro-scale force on microscopic particles and manipulating three-dimensional (3-D) positions of particles. Optical tweezers employ a tightly-focused laser whose beam diameter is smaller than one micrometer (1/100 of hair thickness), which generates attractive force on neighboring microscopic particles moving toward the beam focus. Controlling the positions of the beam focus enabled researchers to hold the particles and move them freely to other locations so they coined the name “optical tweezers.”
To locate the optically-trapped particles by a laser beam, optical microscopes have usually been employed. Optical microscopes measure light signals scattered by the optically-trapped microscopic particles and the positions of the particles in two dimensions. However, it was difficult to quantify the particles’ precise positions along the optic axis, the direction of the beam, from a single image, which is analogous to the difficulty of determining the front and rear positions of objects when closing an eye due to a lack of depth perception. Furthermore, it became more difficult to measure precisely 3-D positions of particles when scattered light signals were distorted by optically-trapped particles having complicated shapes or other particles occlude the target object along the optic axis.
Professor YongKeun Park and his research team in the Department of Physics at the Korea Advanced Institute of Science and Technology (KAIST) employed an optical diffraction tomography (ODT) technique to measure 3-D positions of optically-trapped particles in high speed. The principle of ODT is similar to X-ray CT imaging commonly used in hospitals for visualizing the internal organs of patients. Like X-ray CT imaging, which takes several images from various illumination angles, ODT measures 3-D images of optically-trapped particles by illuminating them with a laser beam in various incidence angles.
The KAIST team used optical tweezers to trap a glass bead with a diameter of 2 micrometers, and moved the bead toward a white blood cell having complicated internal structures. The team measured the 3-D dynamics of the white blood cell as it responded to an approaching glass bead via ODT in the high acquisition rate of 60 images per second. Since the white blood cell screens the glass bead along an optic axis, a conventionally-used optical microscope could not determine the 3-D positions of the glass bead. In contrast, the present method employing ODT localized the 3-D positions of the bead precisely as well as measured the composition of the internal materials of the bead and the white blood cell simultaneously.
Professor Park said, “Our technique has the advantage of measuring the 3-D positions and internal structures of optically-trapped particles in high speed without labelling exogenous fluorescent agents and can be applied in various fields including physics, optics, nanotechnology, and medical science.”
Kyoohyun Kim, the lead author of this paper (“Simultaneous 3D Visualization and Position Tracking of Optically Trapped Particles Using Optical Diffraction Tomography”), added, “This ODT technique can also apply to cellular-level surgeries where optical tweezers are used to manipulate intracellular organelles and to display in real time and in 3-D the images of the reaction of the cell membrane and nucleus during the operation or monitoring the recovery process of the cells from the surgery.”
The research results were published as the cover article in the April 2014 issue of Optica, the newest journal launched last year by the Optical Society of America (OSA) for rapid dissemination of high-impact results related to optics.
Figure 1: This picture shows the concept image of tweezing an optically-trapped glass bead on the cellular membrane of a white blood cell.
Figure 2: High-speed 3-D images produced from optical diffraction tomography technique
2015.04.24 View 13173 -
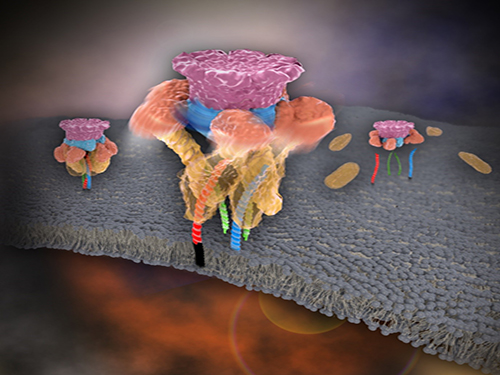 Mystery in Membrane Traffic How NSF Disassembles Single SNAR Complex Solved
KAIST researchers discovered that the protein N-ethylmaleimide-sensitive factor (NSF) unravels a single SNARE complex using one round ATP turnover by tearing the complex with a single burst, contradicting a previous theory that it unwinds in a processive manner.
In 2013, James E. Rothman, Randy W. Schekman, and Thomas C. Südhof won the Nobel Prize in Physiology or Medicine for their discoveries of molecular machineries for vesicle trafficking, a major transport system in cells for maintaining cellular processes. Vesicle traffic acts as a kind of “home-delivery service” in cells. Vesicles package and deliver materials such as proteins and hormones from one cell organelle to another. Then it releases its contents by fusing with the target organelle’s membrane. One example of vesicle traffic is in neuronal communications, where neurotransmitters are released from a neuron. Some of the key proteins for vesicle traffic discovered by the Nobel Prize winners were N-ethylmaleimide-sensitive factor (NSF), alpha-soluble NSF attachment protein (α-SNAP), and soluble SNAP receptors (SNAREs).
SNARE proteins are known as the minimal machinery for membrane fusion. To induce membrane fusion, the proteins combine to form a SNARE complex in a four helical bundle, and NSF and α-SNAP disassemble the SNARE complex for reuse. In particular, NSF can bind an energy source molecule, adenosine triphosphate (ATP), and the ATP-bound NSF develops internal tension via cleavage of ATP. This process is used to exert great force on SNARE complexes, eventually pulling them apart. However, although about 30 years have passed since the Nobel Prize winners’ discovery, how NSF/α-SNAP disassembled the SNARE complex remained a mystery to scientists due to a lack in methodology.
In a recent issue of Science, published on March 27, 2015, a research team, led by Tae-Young Yoon of the Department of Physics at the Korea Advanced Institute of Science and Technology (KAIST) and Reinhard Jahn of the Department of Neurobiology of the Max-Planck-Institute for Biophysical Chemistry, reports that NSF/α-SNAP disassemble a single SNARE complex using various single-molecule biophysical methods that allow them to monitor and manipulate individual protein complexes.
“We have learned that NSF releases energy in a burst within 20 milliseconds to “tear” the SNARE complex apart in a one-step global unfolding reaction, which is immediately followed by the release of SNARE proteins,” said Yoon.
Previously, it was believed that NSF disassembled a SNARE complex by unwinding it in a processive manner. Also, largely unexplained was how many cycles of ATP hydrolysis were required and how these cycles were connected to the disassembly of the SNARE complex.
Yoon added, “From our research, we found that NSF requires hydrolysis of ATPs that were already bound before it attached to the SNAREs—which means that only one round of an ATP turnover is sufficient for SNARE complex disassembly. Moreover, this is possible because NSF pulls a SNARE complex apart by building up the energy from individual ATPs and releasing it at once, yielding a “spring-loaded” mechanism.”
NSF is a member of the ATPases associated with various cellular activities family (AAA+ ATPase), which is essential for many cellular functions such as DNA replication and protein degradation, membrane fusion, microtubule severing, peroxisome biogenesis, signal transduction, and the regulation of gene expression. This research has added valuable new insights and hints for studying AAA+ ATPase proteins, which are crucial for various living beings.
The title of the research paper is “Spring-loaded unraveling of a single SNARE complex by NSF in one round of ATP turnover.” (DOI: 10.1126/science.aaa5267)
Youtube Link: https://www.youtube.com/watch?v=FqTSYHtyHWE&feature=youtu.be
Picture 1. Working model of how NSF/α-SNAP disassemble a single SNARE complex
Picture 2. After neurotransmitter release, NSF disassembles a single SNARE complex using a single round of ATP turnover in a single burst reaction.
2015.03.28 View 12328
Mystery in Membrane Traffic How NSF Disassembles Single SNAR Complex Solved
KAIST researchers discovered that the protein N-ethylmaleimide-sensitive factor (NSF) unravels a single SNARE complex using one round ATP turnover by tearing the complex with a single burst, contradicting a previous theory that it unwinds in a processive manner.
In 2013, James E. Rothman, Randy W. Schekman, and Thomas C. Südhof won the Nobel Prize in Physiology or Medicine for their discoveries of molecular machineries for vesicle trafficking, a major transport system in cells for maintaining cellular processes. Vesicle traffic acts as a kind of “home-delivery service” in cells. Vesicles package and deliver materials such as proteins and hormones from one cell organelle to another. Then it releases its contents by fusing with the target organelle’s membrane. One example of vesicle traffic is in neuronal communications, where neurotransmitters are released from a neuron. Some of the key proteins for vesicle traffic discovered by the Nobel Prize winners were N-ethylmaleimide-sensitive factor (NSF), alpha-soluble NSF attachment protein (α-SNAP), and soluble SNAP receptors (SNAREs).
SNARE proteins are known as the minimal machinery for membrane fusion. To induce membrane fusion, the proteins combine to form a SNARE complex in a four helical bundle, and NSF and α-SNAP disassemble the SNARE complex for reuse. In particular, NSF can bind an energy source molecule, adenosine triphosphate (ATP), and the ATP-bound NSF develops internal tension via cleavage of ATP. This process is used to exert great force on SNARE complexes, eventually pulling them apart. However, although about 30 years have passed since the Nobel Prize winners’ discovery, how NSF/α-SNAP disassembled the SNARE complex remained a mystery to scientists due to a lack in methodology.
In a recent issue of Science, published on March 27, 2015, a research team, led by Tae-Young Yoon of the Department of Physics at the Korea Advanced Institute of Science and Technology (KAIST) and Reinhard Jahn of the Department of Neurobiology of the Max-Planck-Institute for Biophysical Chemistry, reports that NSF/α-SNAP disassemble a single SNARE complex using various single-molecule biophysical methods that allow them to monitor and manipulate individual protein complexes.
“We have learned that NSF releases energy in a burst within 20 milliseconds to “tear” the SNARE complex apart in a one-step global unfolding reaction, which is immediately followed by the release of SNARE proteins,” said Yoon.
Previously, it was believed that NSF disassembled a SNARE complex by unwinding it in a processive manner. Also, largely unexplained was how many cycles of ATP hydrolysis were required and how these cycles were connected to the disassembly of the SNARE complex.
Yoon added, “From our research, we found that NSF requires hydrolysis of ATPs that were already bound before it attached to the SNAREs—which means that only one round of an ATP turnover is sufficient for SNARE complex disassembly. Moreover, this is possible because NSF pulls a SNARE complex apart by building up the energy from individual ATPs and releasing it at once, yielding a “spring-loaded” mechanism.”
NSF is a member of the ATPases associated with various cellular activities family (AAA+ ATPase), which is essential for many cellular functions such as DNA replication and protein degradation, membrane fusion, microtubule severing, peroxisome biogenesis, signal transduction, and the regulation of gene expression. This research has added valuable new insights and hints for studying AAA+ ATPase proteins, which are crucial for various living beings.
The title of the research paper is “Spring-loaded unraveling of a single SNARE complex by NSF in one round of ATP turnover.” (DOI: 10.1126/science.aaa5267)
Youtube Link: https://www.youtube.com/watch?v=FqTSYHtyHWE&feature=youtu.be
Picture 1. Working model of how NSF/α-SNAP disassemble a single SNARE complex
Picture 2. After neurotransmitter release, NSF disassembles a single SNARE complex using a single round of ATP turnover in a single burst reaction.
2015.03.28 View 12328 -
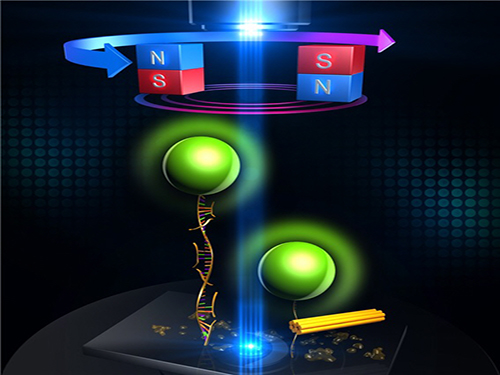 An Advanced Method of DNA Nanostructure Formation Developed
Professor Tae-Young Yoon’s research team from the Department of Physics at KAIST has developed a new method to form DNA nanostructures by using magnetic tweezers to observe and to induce the formation of the structure in real time.
Unlike traditional designs of "DNA origami" which relies on thermal or chemical annealing methods, the new technology utilizes a completely different dynamic in DNA folding. This allows the folding to be done within only ten minutes.
Developed in 2006, the "DNA origami" allows a long skeleton of DNA to be folded into an arbitrary structure by using small stapler DNA pieces. This has been a prominent method in DNA nanotechnology.
However, the traditional technology which adopts thermal processes could not control the DNA formation during the folding because every interaction among DNAs occurs simultaneously. Thus, the thermal processes, which take dozens of hours to complete, had to be repeated multiple times in order to find the optimal condition.
The research team designed a DNA folding using uni-molecular magnetic tweezers that applied force to a single DNA molecule while measuring the state of the DNA. Through this technology, they were able to induce the formation of DNA nanostructure and observe it at the same time.
During high temperature heat treatment, the first stage of conventional thermal processes, the internal structure of the long skeleton DNA untangles. To induce such state, after attaching one side of the skeleton DNA to the surface of glass and the other side to a magnetic material, the team unfolded the internal structure of the DNA by pulling the two sides apart with magnetic force.
Unlike the conventional thermal processes, this method lets the stapler DNA swiftly adhere to the skeleton DNA within a minute because the sites are revealed at room temperature.
After the stapler pieces connected to the skeleton, the team removed the magnetic force. Next, the structure folded through self-assembly as the stapler DNAs stuck to different sites on the skeleton DNA.
Professor Yoon said, “With the existing thermal methods, we could not differentiate the reactions of the DNA because the response of each DNA pieces mutually interacted with each other.” He added that “Using the magnetic tweezers, we were able to sort the process of DNA nanostructure formation into a series of reactions of DNA molecules that are well known, and shorten the time taken for formation in only ten minutes.”
He commented, “This nanostructure formation method will enable us to create more intricate and desirable DNA nanostructures by programming the folding of DNA origami structures.”
Conducted by Dr. Woori Bae under the guidance of Professor Yoon, the research findings were published online in the December 4th issue of Nature Communications.
Figure 1: Uni-molecular magnetic tweezers orchestrating the DNA nanostructure formation
Figure 2: The evolution of DNA nanostructure formation using magnetic tweezers. The DNA nanostructure with a 21-nanometer size was formed in about eight minutes.
2015.01.06 View 8306
An Advanced Method of DNA Nanostructure Formation Developed
Professor Tae-Young Yoon’s research team from the Department of Physics at KAIST has developed a new method to form DNA nanostructures by using magnetic tweezers to observe and to induce the formation of the structure in real time.
Unlike traditional designs of "DNA origami" which relies on thermal or chemical annealing methods, the new technology utilizes a completely different dynamic in DNA folding. This allows the folding to be done within only ten minutes.
Developed in 2006, the "DNA origami" allows a long skeleton of DNA to be folded into an arbitrary structure by using small stapler DNA pieces. This has been a prominent method in DNA nanotechnology.
However, the traditional technology which adopts thermal processes could not control the DNA formation during the folding because every interaction among DNAs occurs simultaneously. Thus, the thermal processes, which take dozens of hours to complete, had to be repeated multiple times in order to find the optimal condition.
The research team designed a DNA folding using uni-molecular magnetic tweezers that applied force to a single DNA molecule while measuring the state of the DNA. Through this technology, they were able to induce the formation of DNA nanostructure and observe it at the same time.
During high temperature heat treatment, the first stage of conventional thermal processes, the internal structure of the long skeleton DNA untangles. To induce such state, after attaching one side of the skeleton DNA to the surface of glass and the other side to a magnetic material, the team unfolded the internal structure of the DNA by pulling the two sides apart with magnetic force.
Unlike the conventional thermal processes, this method lets the stapler DNA swiftly adhere to the skeleton DNA within a minute because the sites are revealed at room temperature.
After the stapler pieces connected to the skeleton, the team removed the magnetic force. Next, the structure folded through self-assembly as the stapler DNAs stuck to different sites on the skeleton DNA.
Professor Yoon said, “With the existing thermal methods, we could not differentiate the reactions of the DNA because the response of each DNA pieces mutually interacted with each other.” He added that “Using the magnetic tweezers, we were able to sort the process of DNA nanostructure formation into a series of reactions of DNA molecules that are well known, and shorten the time taken for formation in only ten minutes.”
He commented, “This nanostructure formation method will enable us to create more intricate and desirable DNA nanostructures by programming the folding of DNA origami structures.”
Conducted by Dr. Woori Bae under the guidance of Professor Yoon, the research findings were published online in the December 4th issue of Nature Communications.
Figure 1: Uni-molecular magnetic tweezers orchestrating the DNA nanostructure formation
Figure 2: The evolution of DNA nanostructure formation using magnetic tweezers. The DNA nanostructure with a 21-nanometer size was formed in about eight minutes.
2015.01.06 View 8306 -
 How Science Understands the Beauty of Fine Arts from the Medieval Era to the 19th Century
A research team, consisting of Professor Hawoong Jeong of the Department of Physics at KAIST and Assistant Professor Seung-Woo Son of the Department of Applied Physics at Hanyang University, conducted a research project to understand visual representations through the eyes of science, i.e., quantitative analyses. Researchers took a sample of reproductions of European paintings from the 11th to the early 19th centuries and analyzed them based on three elements: the usage of color, the variety of painted colors, and the brightness of the images.
For the large-scale quantitative analysis, the research team utilized digital images of the paintings obtained from the Web Gallery of Art, a virtual museum and searchable database of European fine arts that includes over 29,000 pieces, ranging from the years 1000 to 1850. The Web Gallery classifies paintings into ten art historical periods such as Medieval, Renaissance, Mannerist, Baroque, Rococo, Romantic, and Realist.
For each period, researchers investigated the frequency of certain colors which appear in paintings and examined the variety of painted colors, paying particular attention to paintings created by two iconoclastic artists from different eras: Pieter Bruegel the Elder and Jackson Pollock. In their works, the researchers discovered that specific pigments were preferred in each period, the result of reflecting historical facts into fine arts. For example, certain rare colors were used in the medieval age for political and religious reasons, and artists in that era employed a technique to layer one color over another dry color in order to express mixed colors, resulting in thickly textured brushstrokes because they considered mixing colors impure. Moreover, oil colors and color mixing techniques were not fully developed until the Renaissance age.
According to the research team, fewer numbers of colors were used before the 20th century, and the introduction of new expressionist tools, like the use of pastels and fingers directly on canvas, and painting techniques, such as “chiaroscuro” and “sfumato,” made much more colorful and natural expressions possible after the Renaissance period. The team said that the color arrangement of Jackson Pollock’s drip paintings differed substantially from other paintings, showing randomness, especially in the spatial arrangement of colors.
Researchers also examined one of the artistic effects applied to paintings, contrast, an important element to express shape and space in two dimensional fine arts. Among various types of contrasts, they said, brightness contrast is the most important in art history due to the cultural background of Europe which usually adopts the contrast of light and darkness as a metaphorical expression. Taking the color information of pixels and their spatial arrangement, the researchers studied the prevalence of brightness contrast in European paintings over ten artistic periods by developing a correlation function to measure the contrast. These mathematical measurements quantitatively describe the birth of new painting techniques including chiaroscuro and sfumato and their increasing use. For instance, in the medieval age, the contour of objects or images in paintings was vague, but it became much clearer later in the Romantic period.
Professor Jeong said, “The complexity of the material world has been a long-lasting topic of interest in natural science, but research in the structural complexity of art and humanities has only begun since the development of the Internet, with the availability of big data in these fields. Our research is a meaningful attempt to understand the underling intricacy of art and humanities based on a scientific approach, expressed quantitatively.”
The research results were published online on December 11, 2014 in Scientific Reports, entitled “Large-Scale Quantitative Analysis of Painting Arts.” The paper was also selected as one of the weekly research highlights by Nature and is noted on its online journal’s website.
YouTube link on “the brightness contrast”: http://youtu.be/SFo0h1EU2aw
Figure 1: Constructing brightness surfaces and measuring roughness exponents
Figure 2: Visual representations of Mona Lisa painted by Leonardo da Vinci, which was reproduced in accordance with the art historical periods
Figure 3: The screenshot of Nature online webpage
2014.12.23 View 9530
How Science Understands the Beauty of Fine Arts from the Medieval Era to the 19th Century
A research team, consisting of Professor Hawoong Jeong of the Department of Physics at KAIST and Assistant Professor Seung-Woo Son of the Department of Applied Physics at Hanyang University, conducted a research project to understand visual representations through the eyes of science, i.e., quantitative analyses. Researchers took a sample of reproductions of European paintings from the 11th to the early 19th centuries and analyzed them based on three elements: the usage of color, the variety of painted colors, and the brightness of the images.
For the large-scale quantitative analysis, the research team utilized digital images of the paintings obtained from the Web Gallery of Art, a virtual museum and searchable database of European fine arts that includes over 29,000 pieces, ranging from the years 1000 to 1850. The Web Gallery classifies paintings into ten art historical periods such as Medieval, Renaissance, Mannerist, Baroque, Rococo, Romantic, and Realist.
For each period, researchers investigated the frequency of certain colors which appear in paintings and examined the variety of painted colors, paying particular attention to paintings created by two iconoclastic artists from different eras: Pieter Bruegel the Elder and Jackson Pollock. In their works, the researchers discovered that specific pigments were preferred in each period, the result of reflecting historical facts into fine arts. For example, certain rare colors were used in the medieval age for political and religious reasons, and artists in that era employed a technique to layer one color over another dry color in order to express mixed colors, resulting in thickly textured brushstrokes because they considered mixing colors impure. Moreover, oil colors and color mixing techniques were not fully developed until the Renaissance age.
According to the research team, fewer numbers of colors were used before the 20th century, and the introduction of new expressionist tools, like the use of pastels and fingers directly on canvas, and painting techniques, such as “chiaroscuro” and “sfumato,” made much more colorful and natural expressions possible after the Renaissance period. The team said that the color arrangement of Jackson Pollock’s drip paintings differed substantially from other paintings, showing randomness, especially in the spatial arrangement of colors.
Researchers also examined one of the artistic effects applied to paintings, contrast, an important element to express shape and space in two dimensional fine arts. Among various types of contrasts, they said, brightness contrast is the most important in art history due to the cultural background of Europe which usually adopts the contrast of light and darkness as a metaphorical expression. Taking the color information of pixels and their spatial arrangement, the researchers studied the prevalence of brightness contrast in European paintings over ten artistic periods by developing a correlation function to measure the contrast. These mathematical measurements quantitatively describe the birth of new painting techniques including chiaroscuro and sfumato and their increasing use. For instance, in the medieval age, the contour of objects or images in paintings was vague, but it became much clearer later in the Romantic period.
Professor Jeong said, “The complexity of the material world has been a long-lasting topic of interest in natural science, but research in the structural complexity of art and humanities has only begun since the development of the Internet, with the availability of big data in these fields. Our research is a meaningful attempt to understand the underling intricacy of art and humanities based on a scientific approach, expressed quantitatively.”
The research results were published online on December 11, 2014 in Scientific Reports, entitled “Large-Scale Quantitative Analysis of Painting Arts.” The paper was also selected as one of the weekly research highlights by Nature and is noted on its online journal’s website.
YouTube link on “the brightness contrast”: http://youtu.be/SFo0h1EU2aw
Figure 1: Constructing brightness surfaces and measuring roughness exponents
Figure 2: Visual representations of Mona Lisa painted by Leonardo da Vinci, which was reproduced in accordance with the art historical periods
Figure 3: The screenshot of Nature online webpage
2014.12.23 View 9530 -
 Ultra-high Resolution 2-dimentional Real-time Image Capture with Super Lens
Ultra-high Resolution 2-dimentional Real-time Image Capture with Super Lens
Applications to high-precision semiconductor processing or intracellular structures observation are possible.
A joint research team led by Professors Yongkeun Park and Yong-Hoon Cho from the Department of Physics, KAIST, has succeeded in capturing real-time 2D images at a resolution of 100 nm (nanometers), which was impossible with optical lens due to the diffraction limit of light until now. Its future application includes high-precision semiconductor manufacturing process or observation of intracellular structures.
This research follows the past research of the super-lens developed by Professor Park last April, using paint spray to observe images that have three times higher resolution than those discovered by conventional optical lens.
Since optical lens utilize the refraction of light, the diffraction limit, which prevents achieving focus smaller than the wavelength of light, has always been a barrier for acquiring high-resolution images. In the past, it was impossible to observe objects less than the size of 200 to 300 nm in the visible light spectrum.
In order to solve the problem of near-field extinction due to scattering of light, the research team used spray paint consisting of nano-particles massed with dense scattering materials to obtain high-resolution information.
Then, by calculating and restoring the first scattering shape of light using the time reversibility of light, the researchers were able to overcome the diffraction limit. The original position of an object to be observed is obtained by deriving the complex trajectory of the light, and reversing the time to locate the particular position of the object.
Professor Park said, “This new technology can be used as the core technology in all fields which require optical measurement and control. The existing electron microscopy cannot observe cells without destroying them, but the new technology allows us to visualize at ultra-high resolution without destruction.”
The research results were published online in the 9th edition of Physical Review Letters, a prestigious international journal in the field of physics.
2014.09.23 View 11911
Ultra-high Resolution 2-dimentional Real-time Image Capture with Super Lens
Ultra-high Resolution 2-dimentional Real-time Image Capture with Super Lens
Applications to high-precision semiconductor processing or intracellular structures observation are possible.
A joint research team led by Professors Yongkeun Park and Yong-Hoon Cho from the Department of Physics, KAIST, has succeeded in capturing real-time 2D images at a resolution of 100 nm (nanometers), which was impossible with optical lens due to the diffraction limit of light until now. Its future application includes high-precision semiconductor manufacturing process or observation of intracellular structures.
This research follows the past research of the super-lens developed by Professor Park last April, using paint spray to observe images that have three times higher resolution than those discovered by conventional optical lens.
Since optical lens utilize the refraction of light, the diffraction limit, which prevents achieving focus smaller than the wavelength of light, has always been a barrier for acquiring high-resolution images. In the past, it was impossible to observe objects less than the size of 200 to 300 nm in the visible light spectrum.
In order to solve the problem of near-field extinction due to scattering of light, the research team used spray paint consisting of nano-particles massed with dense scattering materials to obtain high-resolution information.
Then, by calculating and restoring the first scattering shape of light using the time reversibility of light, the researchers were able to overcome the diffraction limit. The original position of an object to be observed is obtained by deriving the complex trajectory of the light, and reversing the time to locate the particular position of the object.
Professor Park said, “This new technology can be used as the core technology in all fields which require optical measurement and control. The existing electron microscopy cannot observe cells without destroying them, but the new technology allows us to visualize at ultra-high resolution without destruction.”
The research results were published online in the 9th edition of Physical Review Letters, a prestigious international journal in the field of physics.
2014.09.23 View 11911 -
 Development of a Photonic Diode with Light Speed, Single-Direction Transfer
A photonic diode using a nitride semiconductor rod can increase the possibility of developing all-optical integrated circuits, an alternative to conventional integrated circuits.
Professor Yong-Hoon Cho's research team from the Department of Physics, KAIST, developed a photonic diode which can selectively transfer light in one way, using semiconductor rods.
The photonic diode has a diameter of hundreds of nanometers (nm) and a length of few micrometers. This size enables its use in large-scale integration (LSI). The diode’s less sensitivity towards polarized light angle makes it more useful.
In an integrated circuit, a diode controls the flow of electrons. If this diode controls light rather than electrons, data can be transferred at high speed, and its loss is minimized to a greater extent. Since these implementations conserve more energy, this is a very promising future technology.
However, conventional electronic diodes, made up of asymmetric meta-materials or photonic crystalline structures, are large, which makes them difficult to be used in LSI. These diodes could only be implemented under limited conditions due to its sensitivity towards polarized light angle.
The research team used nitride semiconductor rods to develop a highly efficient photonic diode with distinct light intensities from opposite ends.
The semiconductor rod yields different amount of energy horizontally. According to the research team, this is because the width of the quantum well and its indium quantity is continuously controlled.
Professor Cho said, "A large energy difference in a horizontal direction causes asymmetrical light propagation, enabling it to be operated as a photonic diode." He added that “If light, instead of electrons, were adopted in integrated circuits, the transfer speed would be expected as great as that of light.”
The research findings were published in the September 10th issue of Nano Letters as the cover paper.
Under the guidance of Professor Cho, two Ph.D. candidates, Suk-Min Ko and Su-Hyun Gong, conducted this research. This research project was sponsored by the National Research Foundation of Korea and KAIST’s EEWS (energy, environment, water, and sustainability) Research Center.
Figure Description: Computer simulated image of photonic diode made of semiconductor rod implemented in an all-optical integrated circuit
2014.09.23 View 13357
Development of a Photonic Diode with Light Speed, Single-Direction Transfer
A photonic diode using a nitride semiconductor rod can increase the possibility of developing all-optical integrated circuits, an alternative to conventional integrated circuits.
Professor Yong-Hoon Cho's research team from the Department of Physics, KAIST, developed a photonic diode which can selectively transfer light in one way, using semiconductor rods.
The photonic diode has a diameter of hundreds of nanometers (nm) and a length of few micrometers. This size enables its use in large-scale integration (LSI). The diode’s less sensitivity towards polarized light angle makes it more useful.
In an integrated circuit, a diode controls the flow of electrons. If this diode controls light rather than electrons, data can be transferred at high speed, and its loss is minimized to a greater extent. Since these implementations conserve more energy, this is a very promising future technology.
However, conventional electronic diodes, made up of asymmetric meta-materials or photonic crystalline structures, are large, which makes them difficult to be used in LSI. These diodes could only be implemented under limited conditions due to its sensitivity towards polarized light angle.
The research team used nitride semiconductor rods to develop a highly efficient photonic diode with distinct light intensities from opposite ends.
The semiconductor rod yields different amount of energy horizontally. According to the research team, this is because the width of the quantum well and its indium quantity is continuously controlled.
Professor Cho said, "A large energy difference in a horizontal direction causes asymmetrical light propagation, enabling it to be operated as a photonic diode." He added that “If light, instead of electrons, were adopted in integrated circuits, the transfer speed would be expected as great as that of light.”
The research findings were published in the September 10th issue of Nano Letters as the cover paper.
Under the guidance of Professor Cho, two Ph.D. candidates, Suk-Min Ko and Su-Hyun Gong, conducted this research. This research project was sponsored by the National Research Foundation of Korea and KAIST’s EEWS (energy, environment, water, and sustainability) Research Center.
Figure Description: Computer simulated image of photonic diode made of semiconductor rod implemented in an all-optical integrated circuit
2014.09.23 View 13357 -
 Success in Measuring Protein Interaction at the Molecular Level
Professor Tae Young Yoon
- Live observation of two protein interaction in molecular level successful- The limit in measurement and time resolution of immunoprecipitation technique improved by a hundred thousand fold
KAIST Department of Physics Professor Tae Young Yoon’s research team has successfully observed the interaction of two proteins live on molecular level and the findings were published in the October edition of Nature Protocols.
Professor Yoon’s research team developed a fluorescent microscope that can observe a single molecule. The team grafted the immunoprecipitation technique, traditionally used in protein interaction analysis, to the microscope to develop a “live molecular level immunoprecipitation technique”. The team successfully and accurately measured the reaction between two proteins by repeated momentary interactions in the unit of tens of milliseconds.
The existing immunoprecipitation technique required at least one day to detect interaction between two proteins. There were limitations in detecting momentary or weak interactions. Also, quantitative analysis of the results was difficult since the image was measured by protein-band strength. The technique could not be used for live observation.
The team aimed to drastically improve the existing technique and to develop accurate method of measurement on molecular level. The newly developed technology can enable observation of protein interaction within one hour. Also, the interaction can be measured live, thus the protein interaction phenomenon can be measured in depth.
Moreover, every programme used in the experiment was developed and distributed by the research team so source energy is secured and created the foundation for global infra.
Professor Tae Young Yoon said, “The newly developed technology does not require additional protein expression or purification. Hence, a very small sample of protein is enough to accurately analyse protein interaction on a kinetic level.” He continued, “Even cancerous protein from the tissue of a cancer patient can be analysed. Thus a platform for customised anti-cancer medicine in the future has been prepared, as well.”
Figure 1. Mimetic diagram comparing the existing immunoprecipitation technique and the newly developed live molecular level immunoprecipitation technique
2013.12.11 View 9387
Success in Measuring Protein Interaction at the Molecular Level
Professor Tae Young Yoon
- Live observation of two protein interaction in molecular level successful- The limit in measurement and time resolution of immunoprecipitation technique improved by a hundred thousand fold
KAIST Department of Physics Professor Tae Young Yoon’s research team has successfully observed the interaction of two proteins live on molecular level and the findings were published in the October edition of Nature Protocols.
Professor Yoon’s research team developed a fluorescent microscope that can observe a single molecule. The team grafted the immunoprecipitation technique, traditionally used in protein interaction analysis, to the microscope to develop a “live molecular level immunoprecipitation technique”. The team successfully and accurately measured the reaction between two proteins by repeated momentary interactions in the unit of tens of milliseconds.
The existing immunoprecipitation technique required at least one day to detect interaction between two proteins. There were limitations in detecting momentary or weak interactions. Also, quantitative analysis of the results was difficult since the image was measured by protein-band strength. The technique could not be used for live observation.
The team aimed to drastically improve the existing technique and to develop accurate method of measurement on molecular level. The newly developed technology can enable observation of protein interaction within one hour. Also, the interaction can be measured live, thus the protein interaction phenomenon can be measured in depth.
Moreover, every programme used in the experiment was developed and distributed by the research team so source energy is secured and created the foundation for global infra.
Professor Tae Young Yoon said, “The newly developed technology does not require additional protein expression or purification. Hence, a very small sample of protein is enough to accurately analyse protein interaction on a kinetic level.” He continued, “Even cancerous protein from the tissue of a cancer patient can be analysed. Thus a platform for customised anti-cancer medicine in the future has been prepared, as well.”
Figure 1. Mimetic diagram comparing the existing immunoprecipitation technique and the newly developed live molecular level immunoprecipitation technique
2013.12.11 View 9387