bio
-
 Connecting the Dots to Find New Treatments for Breast Cancer
Systems biologists uncovered new ways of cancer cell reprogramming to treat drug-resistant cancers
Scientists at KAIST believe they may have found a way to reverse an aggressive, treatment-resistant type of breast cancer into a less dangerous kind that responds well to treatment. The study involved the use of mathematical models to untangle the complex genetic and molecular interactions that occur in the two types of breast cancer, but could be extended to find ways for treating many others. The study’s findings were published in the journal Cancer Research.
Basal-like tumours are the most aggressive type of breast cancer, with the worst prognosis. Chemotherapy is the only available treatment option, but patients experience high recurrence rates. On the other hand, luminal-A breast cancer responds well to drugs that specifically target a receptor on their cell surfaces, called estrogen receptor alpha (ERα).
KAIST systems biologist Kwang-Hyun Cho and colleagues analyzed the complex molecular and genetic interactions of basal-like and luminal-A breast cancers to find out if there might be a way to switch the former to the latter and give patients a better chance to respond to treatment.
To do this, they accessed large amounts of cancer and patient data to understand which genes and molecules are involved in the two types. They then input this data into a mathematical model that represents genes, proteins and molecules as dots and the interactions between them as lines. The model can be used to conduct simulations and see how interactions change when certain genes are turned on or off.
“There have been a tremendous number of studies trying to find therapeutic targets for treating basal-like breast cancer patients,” says Cho. “But clinical trials have failed due to the complex and dynamic nature of cancer. To overcome this issue, we looked at breast cancer cells as a complex network system and implemented a systems biological approach to unravel the underlying mechanisms that would allow us to reprogram basal-like into luminal-A breast cancer cells.”
Using this approach, followed by experimental validation on real breast cancer cells, the team found that turning off two key gene regulators, called BCL11A and HDAC1/2, switched a basal-like cancer signalling pathway into a different one used by luminal-A cancer cells. The switch reprograms the cancer cells and makes them more responsive to drugs that target ERα receptors. However, further tests will be needed to confirm that this also works in animal models and eventually humans.
“Our study demonstrates that the systems biological approach can be useful for identifying novel therapeutic targets,” says Cho.
The researchers are now expanding its breast cancer network model to include all breast cancer subtypes. Their ultimate aim is to identify more drug targets and to understand the mechanisms that could drive drug-resistant cells to turn into drug-sensitive ones.
This work was supported by the National Research Foundation of Korea, the Ministry of Science and ICT, Electronics and Telecommunications Research Institute, and the KAIST Grand Challenge 30 Project.
-Publication Sea R. Choi, Chae Young Hwang, Jonghoon Lee, and Kwang-Hyun Cho, “Network Analysis Identifies Regulators of Basal-like Breast Cancer Reprogramming and Endocrine TherapyVulnerability,” Cancer Research, November 30. (doi:10.1158/0008-5472.CAN-21-0621)
-ProfileProfessor Kwang-Hyun ChoLaboratory for Systems Biology and Bio-Inspired EngineeringDepartment of Bio and Brain EngineeringKAIST
2021.12.07 View 12024
Connecting the Dots to Find New Treatments for Breast Cancer
Systems biologists uncovered new ways of cancer cell reprogramming to treat drug-resistant cancers
Scientists at KAIST believe they may have found a way to reverse an aggressive, treatment-resistant type of breast cancer into a less dangerous kind that responds well to treatment. The study involved the use of mathematical models to untangle the complex genetic and molecular interactions that occur in the two types of breast cancer, but could be extended to find ways for treating many others. The study’s findings were published in the journal Cancer Research.
Basal-like tumours are the most aggressive type of breast cancer, with the worst prognosis. Chemotherapy is the only available treatment option, but patients experience high recurrence rates. On the other hand, luminal-A breast cancer responds well to drugs that specifically target a receptor on their cell surfaces, called estrogen receptor alpha (ERα).
KAIST systems biologist Kwang-Hyun Cho and colleagues analyzed the complex molecular and genetic interactions of basal-like and luminal-A breast cancers to find out if there might be a way to switch the former to the latter and give patients a better chance to respond to treatment.
To do this, they accessed large amounts of cancer and patient data to understand which genes and molecules are involved in the two types. They then input this data into a mathematical model that represents genes, proteins and molecules as dots and the interactions between them as lines. The model can be used to conduct simulations and see how interactions change when certain genes are turned on or off.
“There have been a tremendous number of studies trying to find therapeutic targets for treating basal-like breast cancer patients,” says Cho. “But clinical trials have failed due to the complex and dynamic nature of cancer. To overcome this issue, we looked at breast cancer cells as a complex network system and implemented a systems biological approach to unravel the underlying mechanisms that would allow us to reprogram basal-like into luminal-A breast cancer cells.”
Using this approach, followed by experimental validation on real breast cancer cells, the team found that turning off two key gene regulators, called BCL11A and HDAC1/2, switched a basal-like cancer signalling pathway into a different one used by luminal-A cancer cells. The switch reprograms the cancer cells and makes them more responsive to drugs that target ERα receptors. However, further tests will be needed to confirm that this also works in animal models and eventually humans.
“Our study demonstrates that the systems biological approach can be useful for identifying novel therapeutic targets,” says Cho.
The researchers are now expanding its breast cancer network model to include all breast cancer subtypes. Their ultimate aim is to identify more drug targets and to understand the mechanisms that could drive drug-resistant cells to turn into drug-sensitive ones.
This work was supported by the National Research Foundation of Korea, the Ministry of Science and ICT, Electronics and Telecommunications Research Institute, and the KAIST Grand Challenge 30 Project.
-Publication Sea R. Choi, Chae Young Hwang, Jonghoon Lee, and Kwang-Hyun Cho, “Network Analysis Identifies Regulators of Basal-like Breast Cancer Reprogramming and Endocrine TherapyVulnerability,” Cancer Research, November 30. (doi:10.1158/0008-5472.CAN-21-0621)
-ProfileProfessor Kwang-Hyun ChoLaboratory for Systems Biology and Bio-Inspired EngineeringDepartment of Bio and Brain EngineeringKAIST
2021.12.07 View 12024 -
 Scientists Develop Wireless Networks that Allow Brain Circuits to Be Controlled Remotely through the Internet
Wireless implantable devices and IoT could manipulate the brains of animals from anywhere around the world due to their minimalistic hardware, low setup cost, ease of use, and customizable versatility
A new study shows that researchers can remotely control the brain circuits of numerous animals simultaneously and independently through the internet. The scientists believe this newly developed technology can speed up brain research and various neuroscience studies to uncover basic brain functions as well as the underpinnings of various neuropsychiatric and neurological disorders.
A multidisciplinary team of researchers at KAIST, Washington University in St. Louis, and the University of Colorado, Boulder, created a wireless ecosystem with its own wireless implantable devices and Internet of Things (IoT) infrastructure to enable high-throughput neuroscience experiments over the internet. This innovative technology could enable scientists to manipulate the brains of animals from anywhere around the world. The study was published in the journal Nature Biomedical Engineering on November 25
“This novel technology is highly versatile and adaptive. It can remotely control numerous neural implants and laboratory tools in real-time or in a scheduled way without direct human interactions,” said Professor Jae-Woong Jeong of the School of Electrical Engineering at KAIST and a senior author of the study. “These wireless neural devices and equipment integrated with IoT technology have enormous potential for science and medicine.”
The wireless ecosystem only requires a mini-computer that can be purchased for under $45, which connects to the internet and communicates with wireless multifunctional brain probes or other types of conventional laboratory equipment using IoT control modules. By optimally integrating the versatility and modular construction of both unique IoT hardware and software within a single ecosystem, this wireless technology offers new applications that have not been demonstrated before by a single standalone technology. This includes, but is not limited to minimalistic hardware, global remote access, selective and scheduled experiments, customizable automation, and high-throughput scalability.
“As long as researchers have internet access, they are able to trigger, customize, stop, validate, and store the outcomes of large experiments at any time and from anywhere in the world. They can remotely perform large-scale neuroscience experiments in animals deployed in multiple countries,” said one of the lead authors, Dr. Raza Qazi, a researcher with KAIST and the University of Colorado, Boulder. “The low cost of this system allows it to be easily adopted and can further fuel innovation across many laboratories,” Dr. Qazi added.
One of the significant advantages of this IoT neurotechnology is its ability to be mass deployed across the globe due to its minimalistic hardware, low setup cost, ease of use, and customizable versatility. Scientists across the world can quickly implement this technology within their existing laboratories with minimal budget concerns to achieve globally remote access, scalable experimental automation, or both, thus potentially reducing the time needed to unravel various neuroscientific challenges such as those associated with intractable neurological conditions.
Another senior author on the study, Professor Jordan McCall from the Department of Anesthesiology and Center for Clinical Pharmacology at Washington University in St. Louis, said this technology has the potential to change how basic neuroscience studies are performed. “One of the biggest limitations when trying to understand how the mammalian brain works is that we have to study these functions in unnatural conditions. This technology brings us one step closer to performing important studies without direct human interaction with the study subjects.”
The ability to remotely schedule experiments moves toward automating these types of experiments. Dr. Kyle Parker, an instructor at Washington University in St. Louis and another lead author on the study added, “This experimental automation can potentially help us reduce the number of animals used in biomedical research by reducing the variability introduced by various experimenters. This is especially important given our moral imperative to seek research designs that enable this reduction.”
The researchers believe this wireless technology may open new opportunities for many applications including brain research, pharmaceuticals, and telemedicine to treat diseases in the brain and other organs remotely. This remote automation technology could become even more valuable when many labs need to shut down, such as during the height of the COVID-19 pandemic.
This work was supported by grants from the KAIST Global Singularity Research Program, the National Research Foundation of Korea, the United States National Institute of Health, and Oak Ridge Associated Universities.
-PublicationRaza Qazi, Kyle Parker, Choong Yeon Kim, Jordan McCall, Jae-Woong Jeong et al. “Scalable and modular wireless-network infrastructure for large-scale behavioral neuroscience,” Nature Biomedical Engineering, November 25 2021 (doi.org/10.1038/s41551-021-00814-w)
-ProfileProfessor Jae-Woong JeongBio-Integrated Electronics and Systems LabSchool of Electrical EngineeringKAIST
2021.11.29 View 16823
Scientists Develop Wireless Networks that Allow Brain Circuits to Be Controlled Remotely through the Internet
Wireless implantable devices and IoT could manipulate the brains of animals from anywhere around the world due to their minimalistic hardware, low setup cost, ease of use, and customizable versatility
A new study shows that researchers can remotely control the brain circuits of numerous animals simultaneously and independently through the internet. The scientists believe this newly developed technology can speed up brain research and various neuroscience studies to uncover basic brain functions as well as the underpinnings of various neuropsychiatric and neurological disorders.
A multidisciplinary team of researchers at KAIST, Washington University in St. Louis, and the University of Colorado, Boulder, created a wireless ecosystem with its own wireless implantable devices and Internet of Things (IoT) infrastructure to enable high-throughput neuroscience experiments over the internet. This innovative technology could enable scientists to manipulate the brains of animals from anywhere around the world. The study was published in the journal Nature Biomedical Engineering on November 25
“This novel technology is highly versatile and adaptive. It can remotely control numerous neural implants and laboratory tools in real-time or in a scheduled way without direct human interactions,” said Professor Jae-Woong Jeong of the School of Electrical Engineering at KAIST and a senior author of the study. “These wireless neural devices and equipment integrated with IoT technology have enormous potential for science and medicine.”
The wireless ecosystem only requires a mini-computer that can be purchased for under $45, which connects to the internet and communicates with wireless multifunctional brain probes or other types of conventional laboratory equipment using IoT control modules. By optimally integrating the versatility and modular construction of both unique IoT hardware and software within a single ecosystem, this wireless technology offers new applications that have not been demonstrated before by a single standalone technology. This includes, but is not limited to minimalistic hardware, global remote access, selective and scheduled experiments, customizable automation, and high-throughput scalability.
“As long as researchers have internet access, they are able to trigger, customize, stop, validate, and store the outcomes of large experiments at any time and from anywhere in the world. They can remotely perform large-scale neuroscience experiments in animals deployed in multiple countries,” said one of the lead authors, Dr. Raza Qazi, a researcher with KAIST and the University of Colorado, Boulder. “The low cost of this system allows it to be easily adopted and can further fuel innovation across many laboratories,” Dr. Qazi added.
One of the significant advantages of this IoT neurotechnology is its ability to be mass deployed across the globe due to its minimalistic hardware, low setup cost, ease of use, and customizable versatility. Scientists across the world can quickly implement this technology within their existing laboratories with minimal budget concerns to achieve globally remote access, scalable experimental automation, or both, thus potentially reducing the time needed to unravel various neuroscientific challenges such as those associated with intractable neurological conditions.
Another senior author on the study, Professor Jordan McCall from the Department of Anesthesiology and Center for Clinical Pharmacology at Washington University in St. Louis, said this technology has the potential to change how basic neuroscience studies are performed. “One of the biggest limitations when trying to understand how the mammalian brain works is that we have to study these functions in unnatural conditions. This technology brings us one step closer to performing important studies without direct human interaction with the study subjects.”
The ability to remotely schedule experiments moves toward automating these types of experiments. Dr. Kyle Parker, an instructor at Washington University in St. Louis and another lead author on the study added, “This experimental automation can potentially help us reduce the number of animals used in biomedical research by reducing the variability introduced by various experimenters. This is especially important given our moral imperative to seek research designs that enable this reduction.”
The researchers believe this wireless technology may open new opportunities for many applications including brain research, pharmaceuticals, and telemedicine to treat diseases in the brain and other organs remotely. This remote automation technology could become even more valuable when many labs need to shut down, such as during the height of the COVID-19 pandemic.
This work was supported by grants from the KAIST Global Singularity Research Program, the National Research Foundation of Korea, the United States National Institute of Health, and Oak Ridge Associated Universities.
-PublicationRaza Qazi, Kyle Parker, Choong Yeon Kim, Jordan McCall, Jae-Woong Jeong et al. “Scalable and modular wireless-network infrastructure for large-scale behavioral neuroscience,” Nature Biomedical Engineering, November 25 2021 (doi.org/10.1038/s41551-021-00814-w)
-ProfileProfessor Jae-Woong JeongBio-Integrated Electronics and Systems LabSchool of Electrical EngineeringKAIST
2021.11.29 View 16823 -
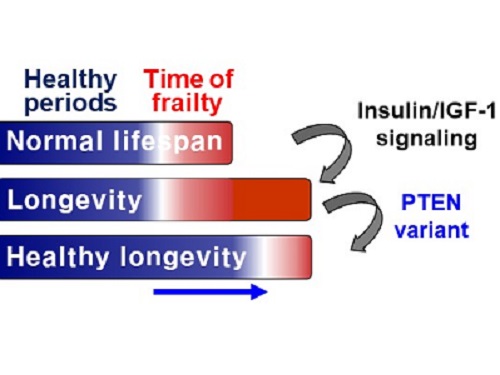 A Genetic Change for Achieving a Long and Healthy Life
Researchers identified a single amino acid change in the tumor suppressor protein in PTEN that extends healthy periods while maintaining longevity
Living a long, healthy life is everyone’s wish, but it is not an easy one to achieve. Many aging studies are developing strategies to increase health spans, the period of life spent with good health, without chronic diseases and disabilities. Researchers at KAIST presented new insights for improving the health span by just regulating the activity of a protein.
A research group under Professor Seung-Jae V. Lee from the Department of Biological Sciences identified a single amino acid change in the tumor suppressor protein phosphatase and tensin homolog (PTEN) that dramatically extends healthy periods while maintaining longevity. This study highlights the importance of the well-conserved tumor suppressor protein PTEN in health span regulation, which can be targeted to develop therapies for promoting healthy longevity in humans. The research was published in Nature Communications on September 24, 2021.
Insulin and insulin-like growth factor-1 (IGF-1) signaling (IIS) is one of the evolutionarily conserved aging-modulatory pathways present in life forms ranging from tiny roundworms to humans. The proper reduction of IIS leads to longevity in animals but often causes defects in multiple health parameters including impaired motility, reproduction, and growth.
The research team found that a specific amino acid change in the PTEN protein improves health status while retaining the longevity conferred by reduced IIS. They used the roundworm C. elegans, an excellent model animal that has been widely used for aging research, mainly because of its very short normal lifespan of about two to three weeks. The PTEN protein is a phosphatase that removes phosphate from lipids as well as proteins. Interestingly, the newly identified amino acid change delicately recalibrated the IIS by partially maintaining protein phosphatase activity while reducing lipid phosphatase activity.
As a result, the amino acid change in the PTEN protein maintained the activity of the longevity-promoting transcription factor Forkhead Box O (FOXO) protein while restricting the detrimental upregulation of another transcription factor, NRF2, leading to long and healthy life in animals with reduced IIS.
Professor Lee said, “Our study raises the exciting possibility of simultaneously promoting longevity and health in humans by slightly tweaking the activity of one protein, PTEN.”
This work was supported by the MInistry of Science and ICT through the National Research Foundation of Korea.
-Publication:Hae-Eun H. Park, Wooseon Hwang, Seokjin Ham, Eunah Kim, Ozlem Altintas, Sangsoon Park, Heehwa G. Son, Yujin Lee, Dongyeop Lee, Won Do Heo, and Seung-Jae V. Lee. 2021. “A PTEN variant uncouples longevity from impaired fitness in Caenorhabditis elegans with reduced insulin/IGF-1 signaling,” Nature Communications, 12(1), 5631. (https://doi.org/10.1038/s41467-021-25920-w)
-ProfileProfessor Seung-Jae V. LeeMolecular Genetics of Aging LaboratoryDepartment of Biological Sciences KAIST
2021.11.19 View 9564
A Genetic Change for Achieving a Long and Healthy Life
Researchers identified a single amino acid change in the tumor suppressor protein in PTEN that extends healthy periods while maintaining longevity
Living a long, healthy life is everyone’s wish, but it is not an easy one to achieve. Many aging studies are developing strategies to increase health spans, the period of life spent with good health, without chronic diseases and disabilities. Researchers at KAIST presented new insights for improving the health span by just regulating the activity of a protein.
A research group under Professor Seung-Jae V. Lee from the Department of Biological Sciences identified a single amino acid change in the tumor suppressor protein phosphatase and tensin homolog (PTEN) that dramatically extends healthy periods while maintaining longevity. This study highlights the importance of the well-conserved tumor suppressor protein PTEN in health span regulation, which can be targeted to develop therapies for promoting healthy longevity in humans. The research was published in Nature Communications on September 24, 2021.
Insulin and insulin-like growth factor-1 (IGF-1) signaling (IIS) is one of the evolutionarily conserved aging-modulatory pathways present in life forms ranging from tiny roundworms to humans. The proper reduction of IIS leads to longevity in animals but often causes defects in multiple health parameters including impaired motility, reproduction, and growth.
The research team found that a specific amino acid change in the PTEN protein improves health status while retaining the longevity conferred by reduced IIS. They used the roundworm C. elegans, an excellent model animal that has been widely used for aging research, mainly because of its very short normal lifespan of about two to three weeks. The PTEN protein is a phosphatase that removes phosphate from lipids as well as proteins. Interestingly, the newly identified amino acid change delicately recalibrated the IIS by partially maintaining protein phosphatase activity while reducing lipid phosphatase activity.
As a result, the amino acid change in the PTEN protein maintained the activity of the longevity-promoting transcription factor Forkhead Box O (FOXO) protein while restricting the detrimental upregulation of another transcription factor, NRF2, leading to long and healthy life in animals with reduced IIS.
Professor Lee said, “Our study raises the exciting possibility of simultaneously promoting longevity and health in humans by slightly tweaking the activity of one protein, PTEN.”
This work was supported by the MInistry of Science and ICT through the National Research Foundation of Korea.
-Publication:Hae-Eun H. Park, Wooseon Hwang, Seokjin Ham, Eunah Kim, Ozlem Altintas, Sangsoon Park, Heehwa G. Son, Yujin Lee, Dongyeop Lee, Won Do Heo, and Seung-Jae V. Lee. 2021. “A PTEN variant uncouples longevity from impaired fitness in Caenorhabditis elegans with reduced insulin/IGF-1 signaling,” Nature Communications, 12(1), 5631. (https://doi.org/10.1038/s41467-021-25920-w)
-ProfileProfessor Seung-Jae V. LeeMolecular Genetics of Aging LaboratoryDepartment of Biological Sciences KAIST
2021.11.19 View 9564 -
 Seegene Opens Covid-19 Testing Mobile Station on Campus
Seegene donates testing reagents for 40,000 people with results available in three hours
Seegene, a molecular diagnostic testing company, donated enough testing reagents for 40,000 COVID-19 tests for the KAIST community and set up a mobile testing station run by the Seegene Medical Foundation on October 28.
The entire COVID-19 diagnosis process, including specimen collection, PCR testing, and results analysis, can be conducted at the mobile testing unit developed by Seegene. The on-site testing station will help the campus get ready to return to normal, especially as the government is transitioning toward its ‘living with Covid-19’ policy, which eases a range of social distancing restrictions. Any KAIST community member can get a Covid-19 test on campus and receive the results within three hours. The station can conduct up to 7,500 tests per day.
This is an extension of the agreement between KAIST and Seegene made in July for research collaboration. The two institutions will work together on various research projects including ultrafast PCR testing, sample collection, and cloud-based data transmission and analysis.
Prior to this donation, according to an administrative order from Daejeon City, KAIST opened a temporary COVID-19 testing center in collaboration with Seegene and conducted COVID-19 tests at the KAIST Clinic over four days starting from September 28. All students living on campus were tested, and all 2,775 tested negative.
Seegene CEO Jong-Yoon Chun said, "KAIST and Seegene signed an agreement for collaborative research on molecular diagnosis in July prior to this donation, and we are happy to maintain a connection with KAIST.” He added, “We hope that this donation will help students return to their ordinary university lives.”
Vice President for Planning and Budget Bowon Kim said, "As KAIST is currently planning to conduct offline lectures in preparation for ‘living with COVID-19’, Seegene’s donation will be particularly helpful.” He added, “The two institutions will continue to cooperate, leading to not only the short-term stabilization of the campus, but also collaborative research for the vitalization of molecular diagnosis technology and the bio industry.”
2021.11.03 View 6864
Seegene Opens Covid-19 Testing Mobile Station on Campus
Seegene donates testing reagents for 40,000 people with results available in three hours
Seegene, a molecular diagnostic testing company, donated enough testing reagents for 40,000 COVID-19 tests for the KAIST community and set up a mobile testing station run by the Seegene Medical Foundation on October 28.
The entire COVID-19 diagnosis process, including specimen collection, PCR testing, and results analysis, can be conducted at the mobile testing unit developed by Seegene. The on-site testing station will help the campus get ready to return to normal, especially as the government is transitioning toward its ‘living with Covid-19’ policy, which eases a range of social distancing restrictions. Any KAIST community member can get a Covid-19 test on campus and receive the results within three hours. The station can conduct up to 7,500 tests per day.
This is an extension of the agreement between KAIST and Seegene made in July for research collaboration. The two institutions will work together on various research projects including ultrafast PCR testing, sample collection, and cloud-based data transmission and analysis.
Prior to this donation, according to an administrative order from Daejeon City, KAIST opened a temporary COVID-19 testing center in collaboration with Seegene and conducted COVID-19 tests at the KAIST Clinic over four days starting from September 28. All students living on campus were tested, and all 2,775 tested negative.
Seegene CEO Jong-Yoon Chun said, "KAIST and Seegene signed an agreement for collaborative research on molecular diagnosis in July prior to this donation, and we are happy to maintain a connection with KAIST.” He added, “We hope that this donation will help students return to their ordinary university lives.”
Vice President for Planning and Budget Bowon Kim said, "As KAIST is currently planning to conduct offline lectures in preparation for ‘living with COVID-19’, Seegene’s donation will be particularly helpful.” He added, “The two institutions will continue to cooperate, leading to not only the short-term stabilization of the campus, but also collaborative research for the vitalization of molecular diagnosis technology and the bio industry.”
2021.11.03 View 6864 -
 New Chiral Nanostructures to Extend the Material Platform
Researchers observed a wide window of chiroptical activity from nanomaterials
A research team transferred chirality from the molecular scale to a microscale to extend material platforms and applications. The optical activity from this novel chiral material encompasses to short-wave infrared region.
This platform could serve as a powerful strategy for hierarchical chirality transfer through self-assembly, generating broad optical activity and providing immense applications including bio, telecommunication, and imaging technique. This is the first observation of such a wide window of chiroptical activity from nanomaterials.
“We synthesized chiral copper sulfides using cysteine, as the stabilizer, and transferring the chirality from molecular to the microscale through self-assembly,” explained Professor Jihyeon Yeom from the Department of Materials Science and Engineering, who led the research. The result was reported in ACS Nano on September 14.
Chiral nanomaterials provide a rich platform for versatile applications. Tuning the wavelength of polarization rotation maxima in the broad range is a promising candidate for infrared neural stimulation, imaging, and nanothermometry. However, the majority of previously developed chiral nanomaterials revealed the optical activity in a relatively shorter wavelength range, not in short-wave infrared.
To achieve chiroptical activity in the short-wave infrared region, materials should be in sub-micrometer dimensions, which are compatible with the wavelength of short-wave infrared region light for strong light-matter interaction. They also should have the optical property of short-wave infrared region absorption while forming a structure with chirality.
Professor Yeom’s team induced self-assembly of the chiral nanoparticles by controlling the attraction and repulsion forces between the building block nanoparticles. During this process, molecular chirality of cysteine was transferred to the nanoscale chirality of nanoparticles, and then transferred to the micrometer scale chirality of nanoflowers with 1.5-2 2 μm dimensions formed by the self-assembly.
“We will work to expand the wavelength range of chiroptical activity to the short-wave infrared region, thus reshaping our daily lives in the form of a bio-barcode that can store vast amount of information under the skin,” said Professor Yeom.
This study was funded by the Ministry of Science and ICT, the Ministry of Health and Welfare, the Ministry of Food and Drug Safety, the National Research Foundation of Korea,the KAIST URP Program, the KAIST Creative Challenging Research Program, Samsung and POSCO Science Fellowship.
-PublicationKi Hyun Park, Junyoung Kwon, Uichang Jeong, Ji-Young Kim, Nicholas A.Kotov, Jihyeon Yeom, “Broad Chrioptical Activity from Ultraviolet to Short-Wave Infrared by Chirality Transfer from Molecular to Micrometer Scale," September 14, 2021 ACS Nano (https://doi.org/10.1021/acsnano.1c05888)
-ProfileProfessor Jihyeon YeomNovel Nanomaterials for New Platforms LaboratoryDepartment of Materials Science and EngineeringKAIST
2021.10.22 View 11363
New Chiral Nanostructures to Extend the Material Platform
Researchers observed a wide window of chiroptical activity from nanomaterials
A research team transferred chirality from the molecular scale to a microscale to extend material platforms and applications. The optical activity from this novel chiral material encompasses to short-wave infrared region.
This platform could serve as a powerful strategy for hierarchical chirality transfer through self-assembly, generating broad optical activity and providing immense applications including bio, telecommunication, and imaging technique. This is the first observation of such a wide window of chiroptical activity from nanomaterials.
“We synthesized chiral copper sulfides using cysteine, as the stabilizer, and transferring the chirality from molecular to the microscale through self-assembly,” explained Professor Jihyeon Yeom from the Department of Materials Science and Engineering, who led the research. The result was reported in ACS Nano on September 14.
Chiral nanomaterials provide a rich platform for versatile applications. Tuning the wavelength of polarization rotation maxima in the broad range is a promising candidate for infrared neural stimulation, imaging, and nanothermometry. However, the majority of previously developed chiral nanomaterials revealed the optical activity in a relatively shorter wavelength range, not in short-wave infrared.
To achieve chiroptical activity in the short-wave infrared region, materials should be in sub-micrometer dimensions, which are compatible with the wavelength of short-wave infrared region light for strong light-matter interaction. They also should have the optical property of short-wave infrared region absorption while forming a structure with chirality.
Professor Yeom’s team induced self-assembly of the chiral nanoparticles by controlling the attraction and repulsion forces between the building block nanoparticles. During this process, molecular chirality of cysteine was transferred to the nanoscale chirality of nanoparticles, and then transferred to the micrometer scale chirality of nanoflowers with 1.5-2 2 μm dimensions formed by the self-assembly.
“We will work to expand the wavelength range of chiroptical activity to the short-wave infrared region, thus reshaping our daily lives in the form of a bio-barcode that can store vast amount of information under the skin,” said Professor Yeom.
This study was funded by the Ministry of Science and ICT, the Ministry of Health and Welfare, the Ministry of Food and Drug Safety, the National Research Foundation of Korea,the KAIST URP Program, the KAIST Creative Challenging Research Program, Samsung and POSCO Science Fellowship.
-PublicationKi Hyun Park, Junyoung Kwon, Uichang Jeong, Ji-Young Kim, Nicholas A.Kotov, Jihyeon Yeom, “Broad Chrioptical Activity from Ultraviolet to Short-Wave Infrared by Chirality Transfer from Molecular to Micrometer Scale," September 14, 2021 ACS Nano (https://doi.org/10.1021/acsnano.1c05888)
-ProfileProfessor Jihyeon YeomNovel Nanomaterials for New Platforms LaboratoryDepartment of Materials Science and EngineeringKAIST
2021.10.22 View 11363 -
 Flexible Sensor-Integrated RFA Needle Leads to Smarter Medical Treatment
Clinical trial of flexible sensor-integrated radiofrequency ablation (RFA) needle tip monitors physical changes and steam pop
Researchers have designed a thin polymeric sensor platform on a radiofrequency ablation needle to monitor temperature and pressure in real time. The sensors integrated onto 1.5 mm diameter needle tip have proven their efficacy during clinical tests and expect to provide a new opportunity for safer and more effective medical practices. The research was reported in Advanced Science as the frontispiece on August 5.
Radiofrequency ablation (RFA) is a minimally invasive surgery technique for removing tumors and treating cardiovascular disease. During a procedure, an unintended audible explosion called ‘steam pop’ can occur due to the increased internal steam pressure in the ablation region. This phenomenon has been cited as a cause of various negative thermal and mechanical effects on neighboring tissue. Even more, the relationship between steam pop and cancer recurrence is still being investigated.
Professor Inkyu Park said that his team’s integrated sensors reliably detected the occurrence of steam pop. The sensors also monitor rapidly spreading hot steam in tissue. It is expected that the diverse properties of tissue undergoing RFA could be checked by utilizing the physical sensors integrated on the needle.
“We believe that the integrated sensors can provide useful information about a variety of medical procedures and accompanying environmental changes in the human body, and help develop more effective and safer surgical procedures,” said Professor Park.
Professor Park’s team built a thin film type pressure and temperature sensor stack with a thickness of less than 10 μm using a microfabrication process. For the pressure sensor, the team used contact resistance changes between metal electrodes and a carbon nanotube coated polymeric membrane. The entire sensor array was thoroughly insulated with medical tubes to minimize any exposure of the sensor materials to external tissue and maximize its biocompatibility.
During the clinical trial, the research team found that the accumulated hot steam is suddenly released during steam pops and this hot air spreads to neighboring tissue, which accelerates the ablation process. Furthermore, using in-situ ultrasound imaging and computational simulations, the research team could confirm the non-uniform temperature distribution around the RFA needle can be one of the primary reasons for the steam popping.
Professor Park explained that various physical and chemical sensors for different targets can be added to create other medical devices and industrial tools.
“This result will expand the usability and applicability of current flexible sensor technologies. We are also trying to integrate this sensor onto a 0.3mm diameter needle for in-vivo diagnosis applications and expect that this approach can be applied to other medical treatments as well as the industrial field,” added Professor Park. This study was supported by the National Research Foundation of Korea.
-PublicationJaeho Park, Jinwoo Lee, Hyo Keun Lim, Inkyu Park et al. “Real-Time Internal Steam Pop Detection during Radiofrequency Ablation with a Radiofrequency Ablation Needle Integrated with a Temperature and Pressure Sensor: Preclinical and clinical pilot tests," Advanced Science (https://doi/org/10.1002/advs.202100725) on August 5, 2021
-ProfileProfessor Inkyu ParkMicro & Nano Tranducers Laboratory http://mintlab1.kaist.ac.kr/
Department of Mechanical EngineeringCollege of EngineeringKAIST
2021.10.20 View 9862
Flexible Sensor-Integrated RFA Needle Leads to Smarter Medical Treatment
Clinical trial of flexible sensor-integrated radiofrequency ablation (RFA) needle tip monitors physical changes and steam pop
Researchers have designed a thin polymeric sensor platform on a radiofrequency ablation needle to monitor temperature and pressure in real time. The sensors integrated onto 1.5 mm diameter needle tip have proven their efficacy during clinical tests and expect to provide a new opportunity for safer and more effective medical practices. The research was reported in Advanced Science as the frontispiece on August 5.
Radiofrequency ablation (RFA) is a minimally invasive surgery technique for removing tumors and treating cardiovascular disease. During a procedure, an unintended audible explosion called ‘steam pop’ can occur due to the increased internal steam pressure in the ablation region. This phenomenon has been cited as a cause of various negative thermal and mechanical effects on neighboring tissue. Even more, the relationship between steam pop and cancer recurrence is still being investigated.
Professor Inkyu Park said that his team’s integrated sensors reliably detected the occurrence of steam pop. The sensors also monitor rapidly spreading hot steam in tissue. It is expected that the diverse properties of tissue undergoing RFA could be checked by utilizing the physical sensors integrated on the needle.
“We believe that the integrated sensors can provide useful information about a variety of medical procedures and accompanying environmental changes in the human body, and help develop more effective and safer surgical procedures,” said Professor Park.
Professor Park’s team built a thin film type pressure and temperature sensor stack with a thickness of less than 10 μm using a microfabrication process. For the pressure sensor, the team used contact resistance changes between metal electrodes and a carbon nanotube coated polymeric membrane. The entire sensor array was thoroughly insulated with medical tubes to minimize any exposure of the sensor materials to external tissue and maximize its biocompatibility.
During the clinical trial, the research team found that the accumulated hot steam is suddenly released during steam pops and this hot air spreads to neighboring tissue, which accelerates the ablation process. Furthermore, using in-situ ultrasound imaging and computational simulations, the research team could confirm the non-uniform temperature distribution around the RFA needle can be one of the primary reasons for the steam popping.
Professor Park explained that various physical and chemical sensors for different targets can be added to create other medical devices and industrial tools.
“This result will expand the usability and applicability of current flexible sensor technologies. We are also trying to integrate this sensor onto a 0.3mm diameter needle for in-vivo diagnosis applications and expect that this approach can be applied to other medical treatments as well as the industrial field,” added Professor Park. This study was supported by the National Research Foundation of Korea.
-PublicationJaeho Park, Jinwoo Lee, Hyo Keun Lim, Inkyu Park et al. “Real-Time Internal Steam Pop Detection during Radiofrequency Ablation with a Radiofrequency Ablation Needle Integrated with a Temperature and Pressure Sensor: Preclinical and clinical pilot tests," Advanced Science (https://doi/org/10.1002/advs.202100725) on August 5, 2021
-ProfileProfessor Inkyu ParkMicro & Nano Tranducers Laboratory http://mintlab1.kaist.ac.kr/
Department of Mechanical EngineeringCollege of EngineeringKAIST
2021.10.20 View 9862 -
 Industrial Liaison Program to Provide Comprehensive Consultation Services
The ILP’s one-stop solutions target all industrial sectors including conglomerates, small and medium-sized enterprises, venture companies, venture capital (VC) firms, and government-affiliated organizations.
The Industrial Liaison Center at KAIST launched the Industrial Liaison Program (ILP) on September 28, an industry-academic cooperation project to provide comprehensive solutions to industry partners. The Industrial Liaison Center will recruit member companies for this service every year, targeting all industrial sectors including conglomerates, small and medium-sized enterprises, venture companies, venture capital (VC) firms, and government-affiliated organizations.
The program plans to build a one-stop support system that can systematically share and use excellent resource information from KAIST’s research teams, R&D achievements, and infrastructure to provide member companies with much-needed services.
More than 40 KAIST professors with abundant academic-industrial collaboration experience will participate in the program. Experts from various fields with different points of view and experiences will jointly provide solutions to ILP member companies. To actively participate in academic-industrial liaisons and joint consultations, KAIST assigned 10 professors from related fields as program directors.
The program directors will come from four different fields including AI/robots (Professor Alice Oh, School from the School of Computing, Professor Young Jae Jang from the Department of Industrial & Systems Engineering, and Professor Yong-Hwa Park from Department of Mechanical Engineering), bio/medicine (Professor Daesoo Kim from Department of Biological Sciences and Professor YongKeun Park from Department of Physics), materials/electronics (Professor Sang Ouk Kim from the Department of Materials Science and Engineering and Professors Jun-Bo Yoon and Seonghwan Cho from the School of Electrical Engineering), and environment/energy (Professor Hee-Tak Kim from the Department of Biological Sciences and Professor Hoon Sohn from the Department of Civil and Environmental Engineering).
The transdisciplinary board of consulting professors that will lead technology innovation is composed of 30 professors including Professor Min-Soo Kim (School of Computing, AI), Professor Chan Hyuk Kim (Department of Biological Sciences, medicine), Professor Hae-Won Park (Department of Mechanical Engineering, robots), Professor Changho Suh (School of Electrical Engineering, electronics), Professor Haeshin Lee (Department of Chemistry, bio), Professor Il-Doo Kim (Department of Materials Science and Engineering, materials), Professor HyeJin Kim (School of Business Technology and Management), and Professor Byoung Pil Kim (School of Business Technology and Management, technology law)
The Head of the Industrial Liaison Center who is also in charge of the program, Professor Keon Jae Lee, said, “In a science and technology-oriented generation where technological supremacy determines national power, it is indispensable to build a new platform upon which innovative academic-industrial cooperation can be pushed forward in the fields of joint consultation, the development of academic-industrial projects, and the foundation of new industries.
He added, “KAIST professors carry out world-class research in many different fields and faculty members can come together through the ILP to communicate with representatives from industry to improve their corporations’ global competitiveness and further contribute to our nation’s interests by cultivating strong small enterprises
2021.09.30 View 7760
Industrial Liaison Program to Provide Comprehensive Consultation Services
The ILP’s one-stop solutions target all industrial sectors including conglomerates, small and medium-sized enterprises, venture companies, venture capital (VC) firms, and government-affiliated organizations.
The Industrial Liaison Center at KAIST launched the Industrial Liaison Program (ILP) on September 28, an industry-academic cooperation project to provide comprehensive solutions to industry partners. The Industrial Liaison Center will recruit member companies for this service every year, targeting all industrial sectors including conglomerates, small and medium-sized enterprises, venture companies, venture capital (VC) firms, and government-affiliated organizations.
The program plans to build a one-stop support system that can systematically share and use excellent resource information from KAIST’s research teams, R&D achievements, and infrastructure to provide member companies with much-needed services.
More than 40 KAIST professors with abundant academic-industrial collaboration experience will participate in the program. Experts from various fields with different points of view and experiences will jointly provide solutions to ILP member companies. To actively participate in academic-industrial liaisons and joint consultations, KAIST assigned 10 professors from related fields as program directors.
The program directors will come from four different fields including AI/robots (Professor Alice Oh, School from the School of Computing, Professor Young Jae Jang from the Department of Industrial & Systems Engineering, and Professor Yong-Hwa Park from Department of Mechanical Engineering), bio/medicine (Professor Daesoo Kim from Department of Biological Sciences and Professor YongKeun Park from Department of Physics), materials/electronics (Professor Sang Ouk Kim from the Department of Materials Science and Engineering and Professors Jun-Bo Yoon and Seonghwan Cho from the School of Electrical Engineering), and environment/energy (Professor Hee-Tak Kim from the Department of Biological Sciences and Professor Hoon Sohn from the Department of Civil and Environmental Engineering).
The transdisciplinary board of consulting professors that will lead technology innovation is composed of 30 professors including Professor Min-Soo Kim (School of Computing, AI), Professor Chan Hyuk Kim (Department of Biological Sciences, medicine), Professor Hae-Won Park (Department of Mechanical Engineering, robots), Professor Changho Suh (School of Electrical Engineering, electronics), Professor Haeshin Lee (Department of Chemistry, bio), Professor Il-Doo Kim (Department of Materials Science and Engineering, materials), Professor HyeJin Kim (School of Business Technology and Management), and Professor Byoung Pil Kim (School of Business Technology and Management, technology law)
The Head of the Industrial Liaison Center who is also in charge of the program, Professor Keon Jae Lee, said, “In a science and technology-oriented generation where technological supremacy determines national power, it is indispensable to build a new platform upon which innovative academic-industrial cooperation can be pushed forward in the fields of joint consultation, the development of academic-industrial projects, and the foundation of new industries.
He added, “KAIST professors carry out world-class research in many different fields and faculty members can come together through the ILP to communicate with representatives from industry to improve their corporations’ global competitiveness and further contribute to our nation’s interests by cultivating strong small enterprises
2021.09.30 View 7760 -
 Two Researchers Designated as SUHF Fellows
Professor Taeyun Ku from the Graduate School of Medical Science and Engineering and Professor Hanseul Yang from the Department of Biological Sciences were nominated as 2021 fellows of the Suh Kyungbae Foundation (SUHF).
SUHF selected three young promising scientists from 53 researchers who are less than five years into their careers. A panel of judges comprised of scholars from home and abroad made the final selection based on the candidates’ innovativeness and power to influence. Professor You-Bong Hyun from Seoul National University also won the fellowship.
Professor Ku’s main topic is opto-connectomics. He will study ways to visualize the complex brain network using innovative technology that transforms neurons into optical elements.
Professor Yang will research the possibility of helping patients recover from skin diseases or injuries without scars by studying spiny mouse genes.
SUHF was established by Amorepacific Group Chairman Suh Kyungbae in 2016 with 300 billion KRW of his private funds. Under the vision of ‘contributing to humanity by supporting innovative discoveries of bioscience researchers,’ the foundation supports promising Korean scientists who pioneer new fields of research in biological sciences.
From 2017 to this year, SUHF has selected 20 promising scientists in the field of biological sciences. Selected scientists are provided with up to KRW 500 million each year for five years. The foundation has provided a total of KRW 48.5 billion in research funds to date.
2021.09.15 View 9709
Two Researchers Designated as SUHF Fellows
Professor Taeyun Ku from the Graduate School of Medical Science and Engineering and Professor Hanseul Yang from the Department of Biological Sciences were nominated as 2021 fellows of the Suh Kyungbae Foundation (SUHF).
SUHF selected three young promising scientists from 53 researchers who are less than five years into their careers. A panel of judges comprised of scholars from home and abroad made the final selection based on the candidates’ innovativeness and power to influence. Professor You-Bong Hyun from Seoul National University also won the fellowship.
Professor Ku’s main topic is opto-connectomics. He will study ways to visualize the complex brain network using innovative technology that transforms neurons into optical elements.
Professor Yang will research the possibility of helping patients recover from skin diseases or injuries without scars by studying spiny mouse genes.
SUHF was established by Amorepacific Group Chairman Suh Kyungbae in 2016 with 300 billion KRW of his private funds. Under the vision of ‘contributing to humanity by supporting innovative discoveries of bioscience researchers,’ the foundation supports promising Korean scientists who pioneer new fields of research in biological sciences.
From 2017 to this year, SUHF has selected 20 promising scientists in the field of biological sciences. Selected scientists are provided with up to KRW 500 million each year for five years. The foundation has provided a total of KRW 48.5 billion in research funds to date.
2021.09.15 View 9709 -
 The Dynamic Tracking of Tissue-Specific Secretory Proteins
Researchers develop a versatile and powerful tool for studying the spatiotemporal dynamics of secretory proteins, a valuable class of biomarkers and therapeutic targets
Researchers have presented a method for profiling tissue-specific secretory proteins in live mice. This method is expected to be applicable to various tissues or disease models for investigating biomarkers or therapeutic targets involved in disease progression. This research was reported in Nature Communications on September 1.
Secretory proteins released into the blood play essential roles in physiological systems. They are core mediators of interorgan communication, while serving as biomarkers and therapeutic targets.
Previous studies have analyzed conditioned media from culture models to identify cell type-specific secretory proteins, but these models often fail to fully recapitulate the intricacies of multi-organ systems and thus do not sufficiently reflect biological realities.
These limitations provided compelling motivation for the research team led by Jae Myoung Suh and his collaborators to develop techniques that could identify and resolve characteristics of tissue-specific secretory proteins along time and space dimensions.
For addressing this gap in the current methodology, the research team utilized proximity-labeling enzymes such as TurboID to label secretory proteins in endoplasmic reticulum lumen using biotin. Thereafter, the biotin-labeled secretory proteins were readily enriched through streptavidin affinity purification and could be identified through mass spectrometry.
To demonstrate its functionality in live mice, research team delivered TurboID to mouse livers via an adenovirus. After administering the biotin, only liver-derived secretory proteins were successfully detected in the plasma of the mice. Interestingly, the pattern of biotin-labeled proteins secreted from the liver was clearly distinctive from those of hepatocyte cell lines.
First author Kwang-eun Kim from the Graduate School of Medical Science and Engineering explained, “The proteins secreted by the liver were significantly different from the results of cell culture models. This data shows the limitations of cell culture models for secretory protein study, and this technique can overcome those limitations. It can be further used to discover biomarkers and therapeutic targets that can more fully reflect the physiological state.”
This work research was supported by the National Research Foundation of Korea, the KAIST Key Research Institutes Project (Interdisciplinary Research Group), and the Institute for Basic Science in Korea.
-PublicationKwang-eun Kim, Isaac Park et al., “Dynamic tracking and identification of tissue-specific secretory proteins in the circulation of live mice,” Nature Communications on Sept.1,
2021(https://doi.org/10.1038/s41467-021-25546-y)
-ProfileProfessor Jae Myoung Suh Integrated Lab of Metabolism, Obesity and Diabetes Researchhttps://imodkaist.wixsite.com/home
Graduate School of Medical Science and Engineering College of Life Science and BioengineeringKAIST
2021.09.14 View 10348
The Dynamic Tracking of Tissue-Specific Secretory Proteins
Researchers develop a versatile and powerful tool for studying the spatiotemporal dynamics of secretory proteins, a valuable class of biomarkers and therapeutic targets
Researchers have presented a method for profiling tissue-specific secretory proteins in live mice. This method is expected to be applicable to various tissues or disease models for investigating biomarkers or therapeutic targets involved in disease progression. This research was reported in Nature Communications on September 1.
Secretory proteins released into the blood play essential roles in physiological systems. They are core mediators of interorgan communication, while serving as biomarkers and therapeutic targets.
Previous studies have analyzed conditioned media from culture models to identify cell type-specific secretory proteins, but these models often fail to fully recapitulate the intricacies of multi-organ systems and thus do not sufficiently reflect biological realities.
These limitations provided compelling motivation for the research team led by Jae Myoung Suh and his collaborators to develop techniques that could identify and resolve characteristics of tissue-specific secretory proteins along time and space dimensions.
For addressing this gap in the current methodology, the research team utilized proximity-labeling enzymes such as TurboID to label secretory proteins in endoplasmic reticulum lumen using biotin. Thereafter, the biotin-labeled secretory proteins were readily enriched through streptavidin affinity purification and could be identified through mass spectrometry.
To demonstrate its functionality in live mice, research team delivered TurboID to mouse livers via an adenovirus. After administering the biotin, only liver-derived secretory proteins were successfully detected in the plasma of the mice. Interestingly, the pattern of biotin-labeled proteins secreted from the liver was clearly distinctive from those of hepatocyte cell lines.
First author Kwang-eun Kim from the Graduate School of Medical Science and Engineering explained, “The proteins secreted by the liver were significantly different from the results of cell culture models. This data shows the limitations of cell culture models for secretory protein study, and this technique can overcome those limitations. It can be further used to discover biomarkers and therapeutic targets that can more fully reflect the physiological state.”
This work research was supported by the National Research Foundation of Korea, the KAIST Key Research Institutes Project (Interdisciplinary Research Group), and the Institute for Basic Science in Korea.
-PublicationKwang-eun Kim, Isaac Park et al., “Dynamic tracking and identification of tissue-specific secretory proteins in the circulation of live mice,” Nature Communications on Sept.1,
2021(https://doi.org/10.1038/s41467-021-25546-y)
-ProfileProfessor Jae Myoung Suh Integrated Lab of Metabolism, Obesity and Diabetes Researchhttps://imodkaist.wixsite.com/home
Graduate School of Medical Science and Engineering College of Life Science and BioengineeringKAIST
2021.09.14 View 10348 -
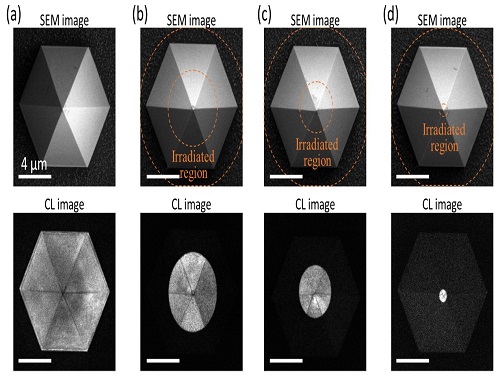 Quantum Emitters: Beyond Crystal Clear to Single-Photon Pure
‘Nanoscale Focus Pinspot’ can quench only the background noise without changing the optical properties of the quantum emitter and the built-in photonic structure
Photons, fundamental particles of light, are carrying these words to your eyes via the light from your computer screen or phone. Photons play a key role in the next-generation quantum information technology, such as quantum computing and communications. A quantum emitter, capable of producing a single, pure photon, is the crux of such technology but has many issues that have yet to be solved, according to KAIST researchers.
A research team under Professor Yong-Hoon Cho has developed a technique that can isolate the desired quality emitter by reducing the noise surrounding the target with what they have dubbed a ‘nanoscale focus pinspot.’ They published their results on June 24 in ACS Nano.
“The nanoscale focus pinspot is a structurally nondestructive technique under an extremely low dose ion beam and is generally applicable for various platforms to improve their single-photon purity while retaining the integrated photonic structures,” said lead author Yong-Hoon Cho from the Department of Physics at KAIST.
To produce single photons from solid state materials, the researchers used wide-bandgap semiconductor quantum dots — fabricated nanoparticles with specialized potential properties, such as the ability to directly inject current into a small chip and to operate at room temperature for practical applications. By making a quantum dot in a photonic structure that propagates light, and then irradiating it with helium ions, researchers theorized that they could develop a quantum emitter that could reduce the unwanted noisy background and produce a single, pure photon on demand.
Professor Cho explained, “Despite its high resolution and versatility, a focused ion beam typically suppresses the optical properties around the bombarded area due to the accelerated ion beam’s high momentum. We focused on the fact that, if the focused ion beam is well controlled, only the background noise can be selectively quenched with high spatial resolution without destroying the structure.”
In other words, the researchers focused the ion beam on a mere pin prick, effectively cutting off the interactions around the quantum dot and removing the physical properties that could negatively interact with and degrade the photon purity emitted from the quantum dot.
“It is the first developed technique that can quench the background noise without changing the optical properties of the quantum emitter and the built-in photonic structure,” Professor Cho asserted.
Professor Cho compared it to stimulated emission depletion microscopy, a technique used to decrease the light around the area of focus, but leaving the focal point illuminated. The result is increased resolution of the desired visual target.
“By adjusting the focused ion beam-irradiated region, we can select the target emitter with nanoscale resolution by quenching the surrounding emitter,” Professor Cho said. “This nanoscale selective-quenching technique can be applied to various material and structural platforms and further extended for applications such as optical memory and high-resolution micro displays.” Korea’s National Research Foundation and the Samsung Science and Technology Foundation supported this work.
-PublicationMinho Choi, Seongmoon Jun, and Yong-Hoon Cho et al. ACS Nano‘Nanoscale Focus Pinspot for High-Purity Quantum Emitters via Focused-Ion-Beam-Induced Luminescence Quenching,’(https://pubs.acs.org/doi/10.1021/acsnano.1c00587)
-ProfileProfessor Yong-Hoon ChoQuantum & Nanobio Photonics Laboratoryhttp://qnp.kaist.ac.kr/
Department of PhysicsKAIST
2021.09.02 View 11926
Quantum Emitters: Beyond Crystal Clear to Single-Photon Pure
‘Nanoscale Focus Pinspot’ can quench only the background noise without changing the optical properties of the quantum emitter and the built-in photonic structure
Photons, fundamental particles of light, are carrying these words to your eyes via the light from your computer screen or phone. Photons play a key role in the next-generation quantum information technology, such as quantum computing and communications. A quantum emitter, capable of producing a single, pure photon, is the crux of such technology but has many issues that have yet to be solved, according to KAIST researchers.
A research team under Professor Yong-Hoon Cho has developed a technique that can isolate the desired quality emitter by reducing the noise surrounding the target with what they have dubbed a ‘nanoscale focus pinspot.’ They published their results on June 24 in ACS Nano.
“The nanoscale focus pinspot is a structurally nondestructive technique under an extremely low dose ion beam and is generally applicable for various platforms to improve their single-photon purity while retaining the integrated photonic structures,” said lead author Yong-Hoon Cho from the Department of Physics at KAIST.
To produce single photons from solid state materials, the researchers used wide-bandgap semiconductor quantum dots — fabricated nanoparticles with specialized potential properties, such as the ability to directly inject current into a small chip and to operate at room temperature for practical applications. By making a quantum dot in a photonic structure that propagates light, and then irradiating it with helium ions, researchers theorized that they could develop a quantum emitter that could reduce the unwanted noisy background and produce a single, pure photon on demand.
Professor Cho explained, “Despite its high resolution and versatility, a focused ion beam typically suppresses the optical properties around the bombarded area due to the accelerated ion beam’s high momentum. We focused on the fact that, if the focused ion beam is well controlled, only the background noise can be selectively quenched with high spatial resolution without destroying the structure.”
In other words, the researchers focused the ion beam on a mere pin prick, effectively cutting off the interactions around the quantum dot and removing the physical properties that could negatively interact with and degrade the photon purity emitted from the quantum dot.
“It is the first developed technique that can quench the background noise without changing the optical properties of the quantum emitter and the built-in photonic structure,” Professor Cho asserted.
Professor Cho compared it to stimulated emission depletion microscopy, a technique used to decrease the light around the area of focus, but leaving the focal point illuminated. The result is increased resolution of the desired visual target.
“By adjusting the focused ion beam-irradiated region, we can select the target emitter with nanoscale resolution by quenching the surrounding emitter,” Professor Cho said. “This nanoscale selective-quenching technique can be applied to various material and structural platforms and further extended for applications such as optical memory and high-resolution micro displays.” Korea’s National Research Foundation and the Samsung Science and Technology Foundation supported this work.
-PublicationMinho Choi, Seongmoon Jun, and Yong-Hoon Cho et al. ACS Nano‘Nanoscale Focus Pinspot for High-Purity Quantum Emitters via Focused-Ion-Beam-Induced Luminescence Quenching,’(https://pubs.acs.org/doi/10.1021/acsnano.1c00587)
-ProfileProfessor Yong-Hoon ChoQuantum & Nanobio Photonics Laboratoryhttp://qnp.kaist.ac.kr/
Department of PhysicsKAIST
2021.09.02 View 11926 -
 Genomic Data Reveals New Insights into Human Embryonic Development
KAIST researchers have used whole-genome sequencing to track the development from a single fertilized-egg to a human body
Genomic scientists at KAIST have revealed new insights into the process of human embryonic development using large-scale, whole-genome sequencing of cells and tissues from adult humans. The study, published in Nature on Aug.25, is the first to analyse somatic mutations in normal tissue across multiple organs within and between humans.
An adult human body comprises trillions of cells of more than 200 types. How a human develops from a single fertilized egg to a fully grown adult is a fundamental question in biomedical science. Due to the ethical challenges of performing studies on human embryos, however, the details of this process remain largely unknown.
To overcome these issues, the research team took a different approach. They analysed genetic mutations in cells taken from adult human post-mortem tissue. Specifically, they identified mutations that occur spontaneously in early developmental cell divisions. These mutations, also called genomic scars, act like unique genetic fingerprints that can be used to trace the embryonic development process.
The study, which looked at 334 single-cell colonies and 379 tissue samples from seven recently deceased human body donors, is the largest single-cell, whole-genome analysis carried out to date. The researchers examined the genomic scars of each individual in order to reconstruct their early embryonic cellular dynamics.
The result revealed several key characteristics of the human embryonic development process. Firstly, mutation rates are higher in the first cell division, but then decrease to approximately one mutation per cell during later cell division. Secondly, early cells contributed unequally to the development of the embryo in all informative donors, for example, at the two-cell stage, one of the cells always left more progeny cells than the other. The ratio of this was different from person to person, implying that the process varies between individuals and is not fully deterministic.
The researchers were also able to deduce the timing of when cells begin to differentiate into individual organ-specific cells. They found that within three days of fertilization, embryonic cells began to be distributed asymmetrically into tissues for the left and right sides of the body, followed by differentiation into three germ layers, and then differentiation into specific tissues and organs.
“It is an impressive scientific achievement that, within 20 years of the completion of human genome project, genomic technology has advanced to the extent that we are now able to accurately identify mutations in a single-cell genome,” said Professor Young Seok Ju from the Graduate School of Medical Science and Engineering at KAIST. “This technology will enable us to track human embryogenesis at even higher resolutions in the future.”
The techniques used in this study could be used to improve our understanding of rare diseases caused by abnormalities in embryonic development, and to design new precision diagnostics and treatments for patients.
The research was completed in collaboration with Kyungpook National University Hospital, the Korea Institute of Science and Technology Information, Catholic University of Korea School of Medicine, Genome Insights Inc, and Immune Square Inc. This work was supported by the Suh Kyungbae Foundation, the Ministry of Health and Welfare of Korea, the National Research Foundastion of Korea.
-PublicationSeongyeol Park, Nanda Mali, Ryul Kim et al. ‘Clonal dynamics in early human embryogenesis inferred from somatic mutation’ Nature Online ahead of print, Aug. 25, 2021 (https://doi.org/10.1038/s41586-021-03786-8)
-ProfileProfessor Young Seok JuLab of Cancer Genomics (https://www.julab.kaist.ac.kr/)Graduate School of Medical Science and EngineeringKAIST
2021.08.31 View 10100
Genomic Data Reveals New Insights into Human Embryonic Development
KAIST researchers have used whole-genome sequencing to track the development from a single fertilized-egg to a human body
Genomic scientists at KAIST have revealed new insights into the process of human embryonic development using large-scale, whole-genome sequencing of cells and tissues from adult humans. The study, published in Nature on Aug.25, is the first to analyse somatic mutations in normal tissue across multiple organs within and between humans.
An adult human body comprises trillions of cells of more than 200 types. How a human develops from a single fertilized egg to a fully grown adult is a fundamental question in biomedical science. Due to the ethical challenges of performing studies on human embryos, however, the details of this process remain largely unknown.
To overcome these issues, the research team took a different approach. They analysed genetic mutations in cells taken from adult human post-mortem tissue. Specifically, they identified mutations that occur spontaneously in early developmental cell divisions. These mutations, also called genomic scars, act like unique genetic fingerprints that can be used to trace the embryonic development process.
The study, which looked at 334 single-cell colonies and 379 tissue samples from seven recently deceased human body donors, is the largest single-cell, whole-genome analysis carried out to date. The researchers examined the genomic scars of each individual in order to reconstruct their early embryonic cellular dynamics.
The result revealed several key characteristics of the human embryonic development process. Firstly, mutation rates are higher in the first cell division, but then decrease to approximately one mutation per cell during later cell division. Secondly, early cells contributed unequally to the development of the embryo in all informative donors, for example, at the two-cell stage, one of the cells always left more progeny cells than the other. The ratio of this was different from person to person, implying that the process varies between individuals and is not fully deterministic.
The researchers were also able to deduce the timing of when cells begin to differentiate into individual organ-specific cells. They found that within three days of fertilization, embryonic cells began to be distributed asymmetrically into tissues for the left and right sides of the body, followed by differentiation into three germ layers, and then differentiation into specific tissues and organs.
“It is an impressive scientific achievement that, within 20 years of the completion of human genome project, genomic technology has advanced to the extent that we are now able to accurately identify mutations in a single-cell genome,” said Professor Young Seok Ju from the Graduate School of Medical Science and Engineering at KAIST. “This technology will enable us to track human embryogenesis at even higher resolutions in the future.”
The techniques used in this study could be used to improve our understanding of rare diseases caused by abnormalities in embryonic development, and to design new precision diagnostics and treatments for patients.
The research was completed in collaboration with Kyungpook National University Hospital, the Korea Institute of Science and Technology Information, Catholic University of Korea School of Medicine, Genome Insights Inc, and Immune Square Inc. This work was supported by the Suh Kyungbae Foundation, the Ministry of Health and Welfare of Korea, the National Research Foundastion of Korea.
-PublicationSeongyeol Park, Nanda Mali, Ryul Kim et al. ‘Clonal dynamics in early human embryogenesis inferred from somatic mutation’ Nature Online ahead of print, Aug. 25, 2021 (https://doi.org/10.1038/s41586-021-03786-8)
-ProfileProfessor Young Seok JuLab of Cancer Genomics (https://www.julab.kaist.ac.kr/)Graduate School of Medical Science and EngineeringKAIST
2021.08.31 View 10100 -
 A Mechanism Underlying Most Common Cause of Epileptic Seizures Revealed
An interdisciplinary study shows that neurons carrying somatic mutations in MTOR can lead to focal epileptogenesis via non-cell-autonomous hyperexcitability of nearby nonmutated neurons
During fetal development, cells should migrate to the outer edge of the brain to form critical connections for information transfer and regulation in the body. When even a few cells fail to move to the correct location, the neurons become disorganized and this results in focal cortical dysplasia. This condition is the most common cause of seizures that cannot be controlled with medication in children and the second most common cause in adults.
Now, an interdisciplinary team studying neurogenetics, neural networks, and neurophysiology at KAIST has revealed how dysfunctions in even a small percentage of cells can cause disorder across the entire brain. They published their results on June 28 in Annals of Neurology.
The work builds on a previous finding, also by a KAIST scientists, who found that focal cortical dysplasia was caused by mutations in the cells involved in mTOR, a pathway that regulates signaling between neurons in the brain.
“Only 1 to 2% of neurons carrying mutations in the mTOR signaling pathway that regulates cell signaling in the brain have been found to include seizures in animal models of focal cortical dysplasia,” said Professor Jong-Woo Sohn from the Department of Biological Sciences. “The main challenge of this study was to explain how nearby non-mutated neurons are hyperexcitable.”
Initially, the researchers hypothesized that the mutated cells affected the number of excitatory and inhibitory synapses in all neurons, mutated or not. These neural gates can trigger or halt activity, respectively, in other neurons. Seizures are a result of extreme activity, called hyperexcitability. If the mutated cells upend the balance and result in more excitatory cells, the researchers thought, it made sense that the cells would be more susceptible to hyperexcitability and, as a result, seizures.
“Contrary to our expectations, the synaptic input balance was not changed in either the mutated or non-mutated neurons,” said Professor Jeong Ho Lee from the Graduate School of Medical Science and Engineering. “We turned our attention to a protein overproduced by mutated neurons.”
The protein is adenosine kinase, which lowers the concentration of adenosine. This naturally occurring compound is an anticonvulsant and works to relax vessels. In mice engineered to have focal cortical dysplasia, the researchers injected adenosine to replace the levels lowered by the protein. It worked and the neurons became less excitable.
“We demonstrated that augmentation of adenosine signaling could attenuate the excitability of non-mutated neurons,” said Professor Se-Bum Paik from the Department of Bio and Brain Engineering.
The effect on the non-mutated neurons was the surprising part, according to Paik. “The seizure-triggering hyperexcitability originated not in the mutation-carrying neurons, but instead in the nearby non-mutated neurons,” he said.
The mutated neurons excreted more adenosine kinase, reducing the adenosine levels in the local environment of all the cells. With less adenosine, the non-mutated neurons became hyperexcitable, leading to seizures.
“While we need further investigate into the relationship between the concentration of adenosine and the increased excitation of nearby neurons, our results support the medical use of drugs to activate adenosine signaling as a possible treatment pathway for focal cortical dysplasia,” Professor Lee said.
The Suh Kyungbae Foundation, the Korea Health Technology Research and Development Project, the Ministry of Health & Welfare, and the National Research Foundation in Korea funded this work.
-Publication:Koh, H.Y., Jang, J., Ju, S.H., Kim, R., Cho, G.-B., Kim, D.S., Sohn, J.-W., Paik, S.-B. and Lee, J.H. (2021), ‘Non–Cell Autonomous Epileptogenesis in Focal Cortical Dysplasia’ Annals of Neurology, 90: 285 299. (https://doi.org/10.1002/ana.26149)
-ProfileProfessor Jeong Ho Lee Translational Neurogenetics Labhttps://tnl.kaist.ac.kr/ Graduate School of Medical Science and Engineering KAIST
Professor Se-Bum Paik Visual System and Neural Network Laboratory http://vs.kaist.ac.kr/ Department of Bio and Brain EngineeringKAIST
Professor Jong-Woo Sohn Laboratory for Neurophysiology, https://sites.google.com/site/sohnlab2014/home Department of Biological SciencesKAIST
Dr. Hyun Yong Koh Translational Neurogenetics LabGraduate School of Medical Science and EngineeringKAIST
Dr. Jaeson Jang Ph.D.Visual System and Neural Network LaboratoryDepartment of Bio and Brain Engineering KAIST
Sang Hyeon Ju M.D.Laboratory for NeurophysiologyDepartment of Biological SciencesKAIST
2021.08.26 View 14756
A Mechanism Underlying Most Common Cause of Epileptic Seizures Revealed
An interdisciplinary study shows that neurons carrying somatic mutations in MTOR can lead to focal epileptogenesis via non-cell-autonomous hyperexcitability of nearby nonmutated neurons
During fetal development, cells should migrate to the outer edge of the brain to form critical connections for information transfer and regulation in the body. When even a few cells fail to move to the correct location, the neurons become disorganized and this results in focal cortical dysplasia. This condition is the most common cause of seizures that cannot be controlled with medication in children and the second most common cause in adults.
Now, an interdisciplinary team studying neurogenetics, neural networks, and neurophysiology at KAIST has revealed how dysfunctions in even a small percentage of cells can cause disorder across the entire brain. They published their results on June 28 in Annals of Neurology.
The work builds on a previous finding, also by a KAIST scientists, who found that focal cortical dysplasia was caused by mutations in the cells involved in mTOR, a pathway that regulates signaling between neurons in the brain.
“Only 1 to 2% of neurons carrying mutations in the mTOR signaling pathway that regulates cell signaling in the brain have been found to include seizures in animal models of focal cortical dysplasia,” said Professor Jong-Woo Sohn from the Department of Biological Sciences. “The main challenge of this study was to explain how nearby non-mutated neurons are hyperexcitable.”
Initially, the researchers hypothesized that the mutated cells affected the number of excitatory and inhibitory synapses in all neurons, mutated or not. These neural gates can trigger or halt activity, respectively, in other neurons. Seizures are a result of extreme activity, called hyperexcitability. If the mutated cells upend the balance and result in more excitatory cells, the researchers thought, it made sense that the cells would be more susceptible to hyperexcitability and, as a result, seizures.
“Contrary to our expectations, the synaptic input balance was not changed in either the mutated or non-mutated neurons,” said Professor Jeong Ho Lee from the Graduate School of Medical Science and Engineering. “We turned our attention to a protein overproduced by mutated neurons.”
The protein is adenosine kinase, which lowers the concentration of adenosine. This naturally occurring compound is an anticonvulsant and works to relax vessels. In mice engineered to have focal cortical dysplasia, the researchers injected adenosine to replace the levels lowered by the protein. It worked and the neurons became less excitable.
“We demonstrated that augmentation of adenosine signaling could attenuate the excitability of non-mutated neurons,” said Professor Se-Bum Paik from the Department of Bio and Brain Engineering.
The effect on the non-mutated neurons was the surprising part, according to Paik. “The seizure-triggering hyperexcitability originated not in the mutation-carrying neurons, but instead in the nearby non-mutated neurons,” he said.
The mutated neurons excreted more adenosine kinase, reducing the adenosine levels in the local environment of all the cells. With less adenosine, the non-mutated neurons became hyperexcitable, leading to seizures.
“While we need further investigate into the relationship between the concentration of adenosine and the increased excitation of nearby neurons, our results support the medical use of drugs to activate adenosine signaling as a possible treatment pathway for focal cortical dysplasia,” Professor Lee said.
The Suh Kyungbae Foundation, the Korea Health Technology Research and Development Project, the Ministry of Health & Welfare, and the National Research Foundation in Korea funded this work.
-Publication:Koh, H.Y., Jang, J., Ju, S.H., Kim, R., Cho, G.-B., Kim, D.S., Sohn, J.-W., Paik, S.-B. and Lee, J.H. (2021), ‘Non–Cell Autonomous Epileptogenesis in Focal Cortical Dysplasia’ Annals of Neurology, 90: 285 299. (https://doi.org/10.1002/ana.26149)
-ProfileProfessor Jeong Ho Lee Translational Neurogenetics Labhttps://tnl.kaist.ac.kr/ Graduate School of Medical Science and Engineering KAIST
Professor Se-Bum Paik Visual System and Neural Network Laboratory http://vs.kaist.ac.kr/ Department of Bio and Brain EngineeringKAIST
Professor Jong-Woo Sohn Laboratory for Neurophysiology, https://sites.google.com/site/sohnlab2014/home Department of Biological SciencesKAIST
Dr. Hyun Yong Koh Translational Neurogenetics LabGraduate School of Medical Science and EngineeringKAIST
Dr. Jaeson Jang Ph.D.Visual System and Neural Network LaboratoryDepartment of Bio and Brain Engineering KAIST
Sang Hyeon Ju M.D.Laboratory for NeurophysiologyDepartment of Biological SciencesKAIST
2021.08.26 View 14756