protein
-
 KAIST Team Develops Optogenetic Platform for Spatiotemporal Control of Protein and mRNA Storage and Release
<Dr. Chaeyeon Lee, Professor Won Do Heo from Department of Biological Sciences>
A KAIST research team led by Professor Won Do Heo (Department of Biological Sciences) has developed an optogenetic platform, RELISR (REversible LIght-induced Store and Release), that enables precise spatiotemporal control over the storage and release of proteins and mRNAs in living cells and animals.
Traditional optogenetic condensate systems have been limited by their reliance on non-specific multivalent interactions, which can lead to unintended sequestration or release of endogenous molecules. RELISR overcomes these limitations by employing highly specific protein–protein (nanobody–antigen) and protein–RNA (MCP–MS2) interactions, enabling the selective and reversible compartmentalization of target proteins or mRNAs within engineered, membrane-less condensates.
In the dark, RELISR stably sequesters target molecules within condensates, physically isolating them from the cellular environment. Upon blue light stimulation, the condensates rapidly dissolve, releasing the stored proteins or mRNAs, which immediately regain their cellular functions or translational competency. This allows for reversible and rapid modulation of molecular activities in response to optical cues.
< Figure 1. Overview of the Artificial Condensate System (RELISR). The artificial condensate system, RELISR, includes "Protein-RELISR" for storing proteins and "mRNA-RELISR" for storing mRNA. These artificial condensates can be disassembled by blue light irradiation and reassembled in a dark state>
The research team demonstrated that RELISR enables temporal and spatial regulation of protein activity and mRNA translation in various cell types, including cultured neurons and mouse liver tissue. Comparative studies showed that RELISR provides more robust and reversible control of translation than previous systems based on spatial translocation.
While previous optogenetic systems such as LARIAT (Lee et al., Nature Methods, 2014) and mRNA-LARIAT (Kim et al., Nat. Cell Biol., 2019) enabled the selective sequestration of proteins or mRNAs into membrane-less condensates in response to light, they were primarily limited to the trapping phase. The RELISR platform introduced in this study establishes a new paradigm by enabling both the targeted storage of proteins and mRNAs and their rapid, light-triggered release. This approach allows researchers to not only confine molecular function on demand, but also to restore activity with precise temporal control.
< Figure 2. Cell shape change using the artificial condensate system (RELISR). A target protein, Vav2, which contributes to cell shape, was stored within the artificial condensate and then released after light irradiation. This release activated the target protein Vav2, causing a change in cell shape. It was confirmed that the storage, release, and activation of various proteins were effectively achieved>
Professor Heo stated, “RELISR is a versatile optogenetic tool that enables the precise control of protein and mRNA function at defined times and locations in living systems. We anticipate this platform will be broadly applicable for studies of cell signaling, neural circuits, and therapeutic development. Furthermore, the combination of RELISR with genome editing or tissue-targeted delivery could further expand its utility for molecular medicine.”
< Figure 3. Expression of a target mRNA using the artificial condensate system (RELISR) in mice. The genetic material for the artificial condensate system, RELISR, was injected into a living mouse. Using this system, a target mRNA was stored within the mouse's liver. Upon light irradiation, the mRNA was released, which induced the translation of a luminescent protein>
This research was conducted by first author Dr. Chaeyeon Lee, under the supervision of Professor Heo, with contributions from Dr. Daseuli Yu (co-corresponding author) and Professor YongKeun Park (co-corresponding author, Department of Physics), whose group performed quantitative imaging analyses of biophysical changes induced by RELISR in cells.
The findings were published in Nature Communications (July 7, 2025; DOI: 10.1038/s41467-025-61322-y). This work was supported by the Samsung Future Technology Foundation and the National Research Foundation of Korea.
2025.07.23 View 275
KAIST Team Develops Optogenetic Platform for Spatiotemporal Control of Protein and mRNA Storage and Release
<Dr. Chaeyeon Lee, Professor Won Do Heo from Department of Biological Sciences>
A KAIST research team led by Professor Won Do Heo (Department of Biological Sciences) has developed an optogenetic platform, RELISR (REversible LIght-induced Store and Release), that enables precise spatiotemporal control over the storage and release of proteins and mRNAs in living cells and animals.
Traditional optogenetic condensate systems have been limited by their reliance on non-specific multivalent interactions, which can lead to unintended sequestration or release of endogenous molecules. RELISR overcomes these limitations by employing highly specific protein–protein (nanobody–antigen) and protein–RNA (MCP–MS2) interactions, enabling the selective and reversible compartmentalization of target proteins or mRNAs within engineered, membrane-less condensates.
In the dark, RELISR stably sequesters target molecules within condensates, physically isolating them from the cellular environment. Upon blue light stimulation, the condensates rapidly dissolve, releasing the stored proteins or mRNAs, which immediately regain their cellular functions or translational competency. This allows for reversible and rapid modulation of molecular activities in response to optical cues.
< Figure 1. Overview of the Artificial Condensate System (RELISR). The artificial condensate system, RELISR, includes "Protein-RELISR" for storing proteins and "mRNA-RELISR" for storing mRNA. These artificial condensates can be disassembled by blue light irradiation and reassembled in a dark state>
The research team demonstrated that RELISR enables temporal and spatial regulation of protein activity and mRNA translation in various cell types, including cultured neurons and mouse liver tissue. Comparative studies showed that RELISR provides more robust and reversible control of translation than previous systems based on spatial translocation.
While previous optogenetic systems such as LARIAT (Lee et al., Nature Methods, 2014) and mRNA-LARIAT (Kim et al., Nat. Cell Biol., 2019) enabled the selective sequestration of proteins or mRNAs into membrane-less condensates in response to light, they were primarily limited to the trapping phase. The RELISR platform introduced in this study establishes a new paradigm by enabling both the targeted storage of proteins and mRNAs and their rapid, light-triggered release. This approach allows researchers to not only confine molecular function on demand, but also to restore activity with precise temporal control.
< Figure 2. Cell shape change using the artificial condensate system (RELISR). A target protein, Vav2, which contributes to cell shape, was stored within the artificial condensate and then released after light irradiation. This release activated the target protein Vav2, causing a change in cell shape. It was confirmed that the storage, release, and activation of various proteins were effectively achieved>
Professor Heo stated, “RELISR is a versatile optogenetic tool that enables the precise control of protein and mRNA function at defined times and locations in living systems. We anticipate this platform will be broadly applicable for studies of cell signaling, neural circuits, and therapeutic development. Furthermore, the combination of RELISR with genome editing or tissue-targeted delivery could further expand its utility for molecular medicine.”
< Figure 3. Expression of a target mRNA using the artificial condensate system (RELISR) in mice. The genetic material for the artificial condensate system, RELISR, was injected into a living mouse. Using this system, a target mRNA was stored within the mouse's liver. Upon light irradiation, the mRNA was released, which induced the translation of a luminescent protein>
This research was conducted by first author Dr. Chaeyeon Lee, under the supervision of Professor Heo, with contributions from Dr. Daseuli Yu (co-corresponding author) and Professor YongKeun Park (co-corresponding author, Department of Physics), whose group performed quantitative imaging analyses of biophysical changes induced by RELISR in cells.
The findings were published in Nature Communications (July 7, 2025; DOI: 10.1038/s41467-025-61322-y). This work was supported by the Samsung Future Technology Foundation and the National Research Foundation of Korea.
2025.07.23 View 275 -
 Why Do Plants Attack Themselves? The Secret of Genetic Conflict Revealed
<Professor Ji-Joon Song of the KAIST Department of Biological Sciences>
Plants, with their unique immune systems, sometimes launch 'autoimmune responses' by mistakenly identifying their own protein structures as pathogens. In particular, 'hybrid necrosis,' a phenomenon where descendant plants fail to grow healthily and perish after cross-breeding different varieties, has long been a difficult challenge for botanists and agricultural researchers. In response, an international research team has successfully elucidated the mechanism inducing plant autoimmune responses and proposed a novel strategy for cultivar improvement that can predict and avoid these reactions.
Professor Ji-Joon Song's research team at KAIST, in collaboration with teams from the National University of Singapore (NUS) and the University of Oxford, announced on the 21st of July that they have elucidated the structure and function of the 'DM3' protein complex, which triggers plant autoimmune responses, using cryo-electron microscopy (Cryo-EM) technology.
This research is drawing attention because it identifies defects in protein structure as the cause of hybrid necrosis, which occurs due to an abnormal reaction of immune receptors during cross-breeding between plant hybrids.
This protein (DM3) is originally an enzyme involved in the plant's immune response, but problems arise when the structure of the DM3 protein is damaged in a specific protein combination called 'DANGEROUS MIX (DM)'.
Notably, one variant of DM3, the 'DM3Col-0' variant, forms a stable complex with six proteins and is recognized as normal, thus not triggering an immune response. In contrast, another 'DM3Hh-0' variant has improper binding between its six proteins, causing the plant to recognize it as an 'abnormal state' and trigger an immune alarm, leading to autoimmunity.
The research team visualized this structure using atomic-resolution cryo-electron microscopy (Cryo-EM) and revealed that the immune-inducing ability is not due to the enzymatic function of the DM3 protein, but rather to 'differences in protein binding affinity.'
<Figure 1. Mechanism of Plant Autoimmunity Triggered by the Collapse of the DM3 Protein Complex>
This demonstrates that plants can initiate an immune response by recognizing not only 'external pathogens' but also 'internal protein structures' when they undergo abnormal changes, treating them as if they were pathogens.
The study shows how sensitively the plant immune system changes and triggers autoimmune responses when genes are mixed and protein structures change during the cross-breeding of different plant varieties. It significantly advanced the understanding of genetic incompatibility that can occur during natural cross-breeding and cultivar improvement processes.
Dr. Gijeong Kim, the co-first author, stated, "Through international research collaboration, we presented a new perspective on understanding the plant immune system by leveraging the autoimmune phenomenon, completing a high-quality study that encompasses structural biochemistry, genetics, and cell biological experiments."
Professor Ji-Joon Song of the KAIST Department of Biological Sciences, who led the research, said, "The fact that the immune system can detect not only external pathogens but also structural abnormalities in its own proteins will set a new standard for plant biotechnology and crop breeding strategies. Cryo-electron microscopy-based structural analysis will be an important tool for understanding the essence of gene interactions."
This research, with Professor Ji-Joon Song and Professor Eunyoung Chae of the University of Oxford as co-corresponding authors, Dr. Gijeong Kim (currently a postdoctoral researcher at the University of Zurich) and Dr. Wei-Lin Wan of the National University of Singapore as co-first authors, and Ph.D candidate Nayun Kim, as the second author, was published on July 17th in Molecular Cell, a sister journal of the international academic journal Cell.
This research was supported by the KAIST Grand Challenge 30 project.
Article Title: Structural determinants of DANGEROUS MIX 3, an alpha/beta hydrolase that triggers NLR-mediated genetic incompatibility in plants DOI: https://doi.org/10.1016/j.molcel.2025.06.021
2025.07.21 View 427
Why Do Plants Attack Themselves? The Secret of Genetic Conflict Revealed
<Professor Ji-Joon Song of the KAIST Department of Biological Sciences>
Plants, with their unique immune systems, sometimes launch 'autoimmune responses' by mistakenly identifying their own protein structures as pathogens. In particular, 'hybrid necrosis,' a phenomenon where descendant plants fail to grow healthily and perish after cross-breeding different varieties, has long been a difficult challenge for botanists and agricultural researchers. In response, an international research team has successfully elucidated the mechanism inducing plant autoimmune responses and proposed a novel strategy for cultivar improvement that can predict and avoid these reactions.
Professor Ji-Joon Song's research team at KAIST, in collaboration with teams from the National University of Singapore (NUS) and the University of Oxford, announced on the 21st of July that they have elucidated the structure and function of the 'DM3' protein complex, which triggers plant autoimmune responses, using cryo-electron microscopy (Cryo-EM) technology.
This research is drawing attention because it identifies defects in protein structure as the cause of hybrid necrosis, which occurs due to an abnormal reaction of immune receptors during cross-breeding between plant hybrids.
This protein (DM3) is originally an enzyme involved in the plant's immune response, but problems arise when the structure of the DM3 protein is damaged in a specific protein combination called 'DANGEROUS MIX (DM)'.
Notably, one variant of DM3, the 'DM3Col-0' variant, forms a stable complex with six proteins and is recognized as normal, thus not triggering an immune response. In contrast, another 'DM3Hh-0' variant has improper binding between its six proteins, causing the plant to recognize it as an 'abnormal state' and trigger an immune alarm, leading to autoimmunity.
The research team visualized this structure using atomic-resolution cryo-electron microscopy (Cryo-EM) and revealed that the immune-inducing ability is not due to the enzymatic function of the DM3 protein, but rather to 'differences in protein binding affinity.'
<Figure 1. Mechanism of Plant Autoimmunity Triggered by the Collapse of the DM3 Protein Complex>
This demonstrates that plants can initiate an immune response by recognizing not only 'external pathogens' but also 'internal protein structures' when they undergo abnormal changes, treating them as if they were pathogens.
The study shows how sensitively the plant immune system changes and triggers autoimmune responses when genes are mixed and protein structures change during the cross-breeding of different plant varieties. It significantly advanced the understanding of genetic incompatibility that can occur during natural cross-breeding and cultivar improvement processes.
Dr. Gijeong Kim, the co-first author, stated, "Through international research collaboration, we presented a new perspective on understanding the plant immune system by leveraging the autoimmune phenomenon, completing a high-quality study that encompasses structural biochemistry, genetics, and cell biological experiments."
Professor Ji-Joon Song of the KAIST Department of Biological Sciences, who led the research, said, "The fact that the immune system can detect not only external pathogens but also structural abnormalities in its own proteins will set a new standard for plant biotechnology and crop breeding strategies. Cryo-electron microscopy-based structural analysis will be an important tool for understanding the essence of gene interactions."
This research, with Professor Ji-Joon Song and Professor Eunyoung Chae of the University of Oxford as co-corresponding authors, Dr. Gijeong Kim (currently a postdoctoral researcher at the University of Zurich) and Dr. Wei-Lin Wan of the National University of Singapore as co-first authors, and Ph.D candidate Nayun Kim, as the second author, was published on July 17th in Molecular Cell, a sister journal of the international academic journal Cell.
This research was supported by the KAIST Grand Challenge 30 project.
Article Title: Structural determinants of DANGEROUS MIX 3, an alpha/beta hydrolase that triggers NLR-mediated genetic incompatibility in plants DOI: https://doi.org/10.1016/j.molcel.2025.06.021
2025.07.21 View 427 -
 KAIST Discovers Protein Switch that Turns Anti-Viral Immune Response On and Off
Even after the COVID-19 pandemic, various new infectious diseases continue to emerge, posing ongoing viral threats that demand robust and sustained immune defenses. However, excessive immune reactions can also harm body tissues, causing significant health issues. KAIST and an international research team have discovered a critical protein that acts as a 'switch' regulating immune responses to viruses. This breakthrough is expected to lay the groundwork for future infectious disease responses and autoimmune disease treatment strategies.
KAIST (President Kwang-Hyung Lee) announced on May 14 that a joint research team led by Professor Yoosik Kim from the Department of Chemical and Biomolecular Engineering at KAIST and Professor Seunghee Cha from University of Florida has discovered the mechanism by which double-stranded RNA derived from mitochondria amplifies immune responses. They identified the protein SLIRP as an 'immune switch' that regulates this process, playing a crucial role in both viral infections and autoimmune diseases.
< (From left) Master's candidate Yewon Yang, Professor Yoosik Kim and Ph.D. candidate Doyeong Ku of the Department of Chemical and Biomolecular Engineering >
Autoimmune diseases arise when the immune system fails to differentiate between external pathogens and the body's own molecules, leading to self-directed attacks. Despite extensive research, the precise causes of excessive inflammatory conditions like Sjögren’s syndrome and systemic lupus erythematosus remain unclear, and effective treatments are still limited.
To uncover the molecular mechanisms driving immune hyperactivation and to identify potential regulatory factors, the research team led by Professor Yoosik Kim focused on mitochondrial double-stranded RNA (mt-dsRNA), a genetic immunogenic material produced within cellular organelles. Since mt-dsRNA structurally resembles viral RNA, it can mistakenly trigger immune responses even in the absence of an actual viral infection.
The team discovered that SLIRP, a key regulator of mt-dsRNA, amplifies immune responses by stabilizing the RNA. They confirmed that SLIRP expression increases in experimental models simulating the tissues of autoimmune disease patients and viral infections. Conversely, suppressing SLIRP significantly reduced the immune response, underscoring its role as a critical factor in immune amplification.
This study also demonstrated the dual function of SLIRP in different contexts. In cells infected with human beta coronavirus OC43 and encephalomyocarditis virus (EMCV), SLIRP suppression led to reduced antiviral responses and increased viral replication. Meanwhile, in the blood and salivary gland cells of Sjögren’s syndrome patients, where both SLIRP and mt-dsRNA levels were elevated, suppressing SLIRP alleviated the abnormal immune response.
These findings highlight SLIRP as a key molecular switch that regulates immune responses in both infections and autoimmune diseases.
< Figure 1. Schematic diagram of antiviral signal amplification by SLIRP: SLIRP-based mt-dsRNA induction, cytoplasmic accumulation, and strong interferon response induction by positive feedback of immune response activation. Confirmation of the immune regulatory function of SLIRP in defense against autoimmune diseases Sjögren's syndrome, coronavirus, and encephalomyocarditis virus infection. >
Professor Yoosik Kim remarked, "Through this study, we have identified SLIRP as a crucial protein that drives immune amplification via mt-dsRNAs. Given its dual role in autoimmune diseases and viral infections, SLIRP presents a promising target for immune regulation therapies across various inflammatory disease contexts."
The study, with Ph.D. student Do-Young Ku (first author) and M.S. student Ye-Won Yang (second author) from the Department of Chemical and Biomolecular Engineering at KAIST as primary contributors, was published online in the journal Cell Reports on April 19, 2025.
※ Paper title: SLIRP amplifies antiviral signaling via positive feedback regulation and contributes to autoimmune diseases※ Main authors: Do-Young Ku (KAIST, first author), Ye-Won Yang (KAIST, second author), Seunghee Cha (University of Florida, corresponding author), Yoosik Kim (KAIST, corresponding author)
This study was supported by the Ministry of Health and Welfare's Public Health Technology Research Program and the National Institutes of Health (NIH) through Research Project (R01) funding.
2025.05.14 View 3928
KAIST Discovers Protein Switch that Turns Anti-Viral Immune Response On and Off
Even after the COVID-19 pandemic, various new infectious diseases continue to emerge, posing ongoing viral threats that demand robust and sustained immune defenses. However, excessive immune reactions can also harm body tissues, causing significant health issues. KAIST and an international research team have discovered a critical protein that acts as a 'switch' regulating immune responses to viruses. This breakthrough is expected to lay the groundwork for future infectious disease responses and autoimmune disease treatment strategies.
KAIST (President Kwang-Hyung Lee) announced on May 14 that a joint research team led by Professor Yoosik Kim from the Department of Chemical and Biomolecular Engineering at KAIST and Professor Seunghee Cha from University of Florida has discovered the mechanism by which double-stranded RNA derived from mitochondria amplifies immune responses. They identified the protein SLIRP as an 'immune switch' that regulates this process, playing a crucial role in both viral infections and autoimmune diseases.
< (From left) Master's candidate Yewon Yang, Professor Yoosik Kim and Ph.D. candidate Doyeong Ku of the Department of Chemical and Biomolecular Engineering >
Autoimmune diseases arise when the immune system fails to differentiate between external pathogens and the body's own molecules, leading to self-directed attacks. Despite extensive research, the precise causes of excessive inflammatory conditions like Sjögren’s syndrome and systemic lupus erythematosus remain unclear, and effective treatments are still limited.
To uncover the molecular mechanisms driving immune hyperactivation and to identify potential regulatory factors, the research team led by Professor Yoosik Kim focused on mitochondrial double-stranded RNA (mt-dsRNA), a genetic immunogenic material produced within cellular organelles. Since mt-dsRNA structurally resembles viral RNA, it can mistakenly trigger immune responses even in the absence of an actual viral infection.
The team discovered that SLIRP, a key regulator of mt-dsRNA, amplifies immune responses by stabilizing the RNA. They confirmed that SLIRP expression increases in experimental models simulating the tissues of autoimmune disease patients and viral infections. Conversely, suppressing SLIRP significantly reduced the immune response, underscoring its role as a critical factor in immune amplification.
This study also demonstrated the dual function of SLIRP in different contexts. In cells infected with human beta coronavirus OC43 and encephalomyocarditis virus (EMCV), SLIRP suppression led to reduced antiviral responses and increased viral replication. Meanwhile, in the blood and salivary gland cells of Sjögren’s syndrome patients, where both SLIRP and mt-dsRNA levels were elevated, suppressing SLIRP alleviated the abnormal immune response.
These findings highlight SLIRP as a key molecular switch that regulates immune responses in both infections and autoimmune diseases.
< Figure 1. Schematic diagram of antiviral signal amplification by SLIRP: SLIRP-based mt-dsRNA induction, cytoplasmic accumulation, and strong interferon response induction by positive feedback of immune response activation. Confirmation of the immune regulatory function of SLIRP in defense against autoimmune diseases Sjögren's syndrome, coronavirus, and encephalomyocarditis virus infection. >
Professor Yoosik Kim remarked, "Through this study, we have identified SLIRP as a crucial protein that drives immune amplification via mt-dsRNAs. Given its dual role in autoimmune diseases and viral infections, SLIRP presents a promising target for immune regulation therapies across various inflammatory disease contexts."
The study, with Ph.D. student Do-Young Ku (first author) and M.S. student Ye-Won Yang (second author) from the Department of Chemical and Biomolecular Engineering at KAIST as primary contributors, was published online in the journal Cell Reports on April 19, 2025.
※ Paper title: SLIRP amplifies antiviral signaling via positive feedback regulation and contributes to autoimmune diseases※ Main authors: Do-Young Ku (KAIST, first author), Ye-Won Yang (KAIST, second author), Seunghee Cha (University of Florida, corresponding author), Yoosik Kim (KAIST, corresponding author)
This study was supported by the Ministry of Health and Welfare's Public Health Technology Research Program and the National Institutes of Health (NIH) through Research Project (R01) funding.
2025.05.14 View 3928 -
 KAIST Accelerates Synthetic Microbe Design by Discovering Novel Enzymes Using AI
< (From left) Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering (top), Hongkeun Ji, PhD candidate of the Department of Chemical and Biomolecular Engineering (top), Ha Rim Kim, PhD candidate of the Department of Chemical and Biomolecular Engineering, and Dr. Gi Bae Kim of the BioProcess Engineering Research Center >
Enzymes are proteins that catalyze biochemical reactions within cells and play a pivotal role in metabolic processes. Accordingly, identifying the functions of novel enzymes is a critical task in the construction of microbial cell factories.
A KAIST research team has leveraged artificial intelligence (AI) to design novel enzymes that do not exist in nature, significantly accelerating microbial cell factory development and boosting the potential for next-generation biotechnological applications such as drug development and biofuel production.
KAIST (represented by President Kwang-Hyung Lee) announced on the 21st of April that Distinguished Professor Sang Yup Lee and his team from the Department of Chemical and Biomolecular Engineering have published a review titled “Enzyme Functional Classification Using Artificial Intelligence,” which outlines the advancement of AI-based enzyme function prediction technologies and analyzes how AI has contributed to the discovery and design of new enzymes.
Professor Lee’s team systematically reviewed the development of enzyme function prediction technologies utilizing machine learning and deep learning, offering a comprehensive analysis.
From sequence similarity-based prediction methods to the integration of convolutional neural networks (CNNs), recurrent neural networks (RNNs), graph neural networks (GNNs), and transformer-based large language models, the paper covers a broad range of AI applications. It analyzes how these technologies extract meaningful information from protein sequences and enhance prediction accuracy.
In particular, enzyme function prediction using deep learning goes beyond simple sequence similarity analysis. By automatically extracting structural and evolutionary features embedded in amino acid sequences, deep learning enables more precise predictions of catalytic functions.
This highlights the unique advantages of AI models compared to traditional bioinformatics approaches.
Moreover, the review suggests that the advancement of generative AI will move future research beyond predicting existing functions to generating entirely new enzymes with functions not found in nature. This shift is expected to profoundly impact the trajectory of biotechnology and synthetic biology.
< Figure 1. Extraction of enzyme characteristics and function prediction using various deep learning structures >
Ha Rim Kim, a Ph.D. candidate and co-first author from the Department of Chemical and Biomolecular Engineering, stated, “AI-based enzyme function prediction and enzyme design are highly important across various fields including metabolic engineering, synthetic biology, and healthcare.”
Distinguished Professor Sang Yup Lee added, “AI-powered enzyme function prediction shows the potential to solve diverse biological problems and will significantly contribute to accelerating research across the entire field.”
The review was published on March 28 in Trends in Biotechnology, a leading biotechnology journal issued by Cell Press.
※ Title: Enzyme Functional Classification Using Artificial Intelligence
※DOI: https://doi.org/10.1016/j.tibtech.2025.03.003
※ Author Information: Ha Rim Kim (KAIST, Co-first author), Hongkeun Ji (KAIST, Co-first author), Gi Bae Kim (KAIST, Third author), Sang Yup Lee (KAIST, Corresponding author)
This research was supported by the Ministry of Science and ICT under the project Development of Core Technologies for Advanced Synthetic Biology to Lead the Bio-Manufacturing Industry (aimed at replacing petroleum-based chemicals), and also by joint support from the Ministry of Science and ICT and the Ministry of Health and Welfare for the project Development of Novel Antibiotic Structures Using Deep Learning-Based Synthetic Biology.
2025.04.07 View 4861
KAIST Accelerates Synthetic Microbe Design by Discovering Novel Enzymes Using AI
< (From left) Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering (top), Hongkeun Ji, PhD candidate of the Department of Chemical and Biomolecular Engineering (top), Ha Rim Kim, PhD candidate of the Department of Chemical and Biomolecular Engineering, and Dr. Gi Bae Kim of the BioProcess Engineering Research Center >
Enzymes are proteins that catalyze biochemical reactions within cells and play a pivotal role in metabolic processes. Accordingly, identifying the functions of novel enzymes is a critical task in the construction of microbial cell factories.
A KAIST research team has leveraged artificial intelligence (AI) to design novel enzymes that do not exist in nature, significantly accelerating microbial cell factory development and boosting the potential for next-generation biotechnological applications such as drug development and biofuel production.
KAIST (represented by President Kwang-Hyung Lee) announced on the 21st of April that Distinguished Professor Sang Yup Lee and his team from the Department of Chemical and Biomolecular Engineering have published a review titled “Enzyme Functional Classification Using Artificial Intelligence,” which outlines the advancement of AI-based enzyme function prediction technologies and analyzes how AI has contributed to the discovery and design of new enzymes.
Professor Lee’s team systematically reviewed the development of enzyme function prediction technologies utilizing machine learning and deep learning, offering a comprehensive analysis.
From sequence similarity-based prediction methods to the integration of convolutional neural networks (CNNs), recurrent neural networks (RNNs), graph neural networks (GNNs), and transformer-based large language models, the paper covers a broad range of AI applications. It analyzes how these technologies extract meaningful information from protein sequences and enhance prediction accuracy.
In particular, enzyme function prediction using deep learning goes beyond simple sequence similarity analysis. By automatically extracting structural and evolutionary features embedded in amino acid sequences, deep learning enables more precise predictions of catalytic functions.
This highlights the unique advantages of AI models compared to traditional bioinformatics approaches.
Moreover, the review suggests that the advancement of generative AI will move future research beyond predicting existing functions to generating entirely new enzymes with functions not found in nature. This shift is expected to profoundly impact the trajectory of biotechnology and synthetic biology.
< Figure 1. Extraction of enzyme characteristics and function prediction using various deep learning structures >
Ha Rim Kim, a Ph.D. candidate and co-first author from the Department of Chemical and Biomolecular Engineering, stated, “AI-based enzyme function prediction and enzyme design are highly important across various fields including metabolic engineering, synthetic biology, and healthcare.”
Distinguished Professor Sang Yup Lee added, “AI-powered enzyme function prediction shows the potential to solve diverse biological problems and will significantly contribute to accelerating research across the entire field.”
The review was published on March 28 in Trends in Biotechnology, a leading biotechnology journal issued by Cell Press.
※ Title: Enzyme Functional Classification Using Artificial Intelligence
※DOI: https://doi.org/10.1016/j.tibtech.2025.03.003
※ Author Information: Ha Rim Kim (KAIST, Co-first author), Hongkeun Ji (KAIST, Co-first author), Gi Bae Kim (KAIST, Third author), Sang Yup Lee (KAIST, Corresponding author)
This research was supported by the Ministry of Science and ICT under the project Development of Core Technologies for Advanced Synthetic Biology to Lead the Bio-Manufacturing Industry (aimed at replacing petroleum-based chemicals), and also by joint support from the Ministry of Science and ICT and the Ministry of Health and Welfare for the project Development of Novel Antibiotic Structures Using Deep Learning-Based Synthetic Biology.
2025.04.07 View 4861 -
 KAIST Captures Protein Reaction in Just Six Milliseconds
Understanding biomolecular processes - such as protein-protein interactions and enzyme-substrate reactions that occur on the microseconds to millisecond time scale is essential for comprehending life processes and advancing drug development. KAIST researchers have developed a method for freezing and analyzing biochemical reaction dynamics within a span of just a few milliseconds, marking a significant step forward in better understanding complex biological reactions.
< Photo. (From left) Professor Jin Young Kang and Haerang Hwang of the Integrated Master's and Doctoral Program of the Department of Chemistry, along with Professor Wonhee Lee of the Department of Physics >
KAIST (represented by President Kwang Hyung Lee) announced on the 24th of March that a joint research team led by Professor Jin Young Kang from the Department of Chemistry and Professor Wonhee Lee from the Department of Physics has developed a parylene-based thin-film microfluidic mixing-and-spraying device for ultra-fast biochemical reaction studies.
*Parylene: A key material for microfluidic devices used to observe protein dynamics at ultra-high speeds. It can be fabricated into a few micrometer-thick films, which can be used in making a spray nozzle for microfluidic devices.
This research overcomes the limitations of the existing time-resolved cryo-electron microscopy (TRCEM) method by reducing sample consumption to one-third of the conventional amount while improving the minimum time resolution—down to just six milliseconds (6 ms).
TRCEM is a technique that rapidly freezes protein complexes during intermediate reaction stages under cryogenic conditions, which allows researchers to analyze their structures. This approach has gained significant attention recently for its ability to capture transient biochemical events.
< Figure 1. Time-resolved cryo-EM (TRCEM) technique using microfluidic channels. In order to capture the intermediate structure of biomolecules during a biochemical reaction over time, biomolecules and reaction substrates are mixed in a microfluidic channel, and then sprayed on a grid after a certain reaction time and frozen in liquid ethane to prepare a cryo-EM sample. This can then be analyzed by cryo-EM to observe the structural changes of proteins over time. >
Transient intermediate structures of protein complexes could not be captured by traditional cryo-electron microscopy due to their extremely short lifespans. Although several TRCEM techniques have been developed to address this issue, previous methods were hindered by large sample consumption and limited time resolution. To overcome these challenges, the KAIST team developed a new mixing-and-spraying device using ultra-thin parylene films. The integrated design of the device further enhanced the precision and reproducibility of experiments.
< Figure 2. TRCEM grid fabrication setup using a parylene-based thin-film microfluidic device and actual appearance of the device. You can see that a thin-film parylene channel is inserted into the injection nozzle. The integration of the reaction channel and the injection nozzle allowed the residence time in the device to be reduced to at least 0.5 ms. >
“This research makes TRCEM more practical and paves the way for diverse applications of the parylene thin-film device in structural biology, drug development, enzyme reaction studies, and biosensor research.” Professor Jin Young Kang explained, emphasizing the significance of the study.
Professor Wonhee Lee added, “The team aims to continue this research, focusing on improvement of the technique to achieve higher time resolution with minimal sample consumption.”
< Figure 3. Comparison of the spraying patterns of the parylene mixing-jet device and the conventional mixing-jet device and the filament length in the resulting RecA-ssDNA filament formation reaction. It was shown that the thin film spray nozzle structure affects the uniformity and accuracy of the final reaction time. >
The research findings, with Haerang Hwang (a graduate student in the integrated master's and Ph.D. program in the Department of Chemistry) as the first author, were published online on January 28, 2025, in the international journal Advanced Functional Materials. (Paper Title: “Integrated Parylene-Based Thin-Film Microfluidic Device for Time-Resolved Cryo-Electron Microscopy”, DOI: doi.org/10.1002/adfm.202418224)
This research was supported by the National Research Foundation of Korea (NRF), the Samsung Future Technology Development Program, and the CELINE consortium.
2025.03.24 View 4985
KAIST Captures Protein Reaction in Just Six Milliseconds
Understanding biomolecular processes - such as protein-protein interactions and enzyme-substrate reactions that occur on the microseconds to millisecond time scale is essential for comprehending life processes and advancing drug development. KAIST researchers have developed a method for freezing and analyzing biochemical reaction dynamics within a span of just a few milliseconds, marking a significant step forward in better understanding complex biological reactions.
< Photo. (From left) Professor Jin Young Kang and Haerang Hwang of the Integrated Master's and Doctoral Program of the Department of Chemistry, along with Professor Wonhee Lee of the Department of Physics >
KAIST (represented by President Kwang Hyung Lee) announced on the 24th of March that a joint research team led by Professor Jin Young Kang from the Department of Chemistry and Professor Wonhee Lee from the Department of Physics has developed a parylene-based thin-film microfluidic mixing-and-spraying device for ultra-fast biochemical reaction studies.
*Parylene: A key material for microfluidic devices used to observe protein dynamics at ultra-high speeds. It can be fabricated into a few micrometer-thick films, which can be used in making a spray nozzle for microfluidic devices.
This research overcomes the limitations of the existing time-resolved cryo-electron microscopy (TRCEM) method by reducing sample consumption to one-third of the conventional amount while improving the minimum time resolution—down to just six milliseconds (6 ms).
TRCEM is a technique that rapidly freezes protein complexes during intermediate reaction stages under cryogenic conditions, which allows researchers to analyze their structures. This approach has gained significant attention recently for its ability to capture transient biochemical events.
< Figure 1. Time-resolved cryo-EM (TRCEM) technique using microfluidic channels. In order to capture the intermediate structure of biomolecules during a biochemical reaction over time, biomolecules and reaction substrates are mixed in a microfluidic channel, and then sprayed on a grid after a certain reaction time and frozen in liquid ethane to prepare a cryo-EM sample. This can then be analyzed by cryo-EM to observe the structural changes of proteins over time. >
Transient intermediate structures of protein complexes could not be captured by traditional cryo-electron microscopy due to their extremely short lifespans. Although several TRCEM techniques have been developed to address this issue, previous methods were hindered by large sample consumption and limited time resolution. To overcome these challenges, the KAIST team developed a new mixing-and-spraying device using ultra-thin parylene films. The integrated design of the device further enhanced the precision and reproducibility of experiments.
< Figure 2. TRCEM grid fabrication setup using a parylene-based thin-film microfluidic device and actual appearance of the device. You can see that a thin-film parylene channel is inserted into the injection nozzle. The integration of the reaction channel and the injection nozzle allowed the residence time in the device to be reduced to at least 0.5 ms. >
“This research makes TRCEM more practical and paves the way for diverse applications of the parylene thin-film device in structural biology, drug development, enzyme reaction studies, and biosensor research.” Professor Jin Young Kang explained, emphasizing the significance of the study.
Professor Wonhee Lee added, “The team aims to continue this research, focusing on improvement of the technique to achieve higher time resolution with minimal sample consumption.”
< Figure 3. Comparison of the spraying patterns of the parylene mixing-jet device and the conventional mixing-jet device and the filament length in the resulting RecA-ssDNA filament formation reaction. It was shown that the thin film spray nozzle structure affects the uniformity and accuracy of the final reaction time. >
The research findings, with Haerang Hwang (a graduate student in the integrated master's and Ph.D. program in the Department of Chemistry) as the first author, were published online on January 28, 2025, in the international journal Advanced Functional Materials. (Paper Title: “Integrated Parylene-Based Thin-Film Microfluidic Device for Time-Resolved Cryo-Electron Microscopy”, DOI: doi.org/10.1002/adfm.202418224)
This research was supported by the National Research Foundation of Korea (NRF), the Samsung Future Technology Development Program, and the CELINE consortium.
2025.03.24 View 4985 -
 KAIST Uncovers the Principles of Gene Expression Regulation in Cancer and Cellular Functions
< (From left) Professor Seyun Kim, Professor Gwangrog Lee, Dr. Hyoungjoon Ahn, Dr. Jeongmin Yu, Professor Won-Ki Cho, and (below) PhD candidate Kwangmin Ryu of the Department of Biological Sciences>
A research team at KAIST has identified the core gene expression networks regulated by key proteins that fundamentally drive phenomena such as cancer development, metastasis, tissue differentiation from stem cells, and neural activation processes. This discovery lays the foundation for developing innovative therapeutic technologies.
On the 22nd of January, KAIST (represented by President Kwang Hyung Lee) announced that the joint research team led by Professors Seyun Kim, Gwangrog Lee, and Won-Ki Cho from the Department of Biological Sciences had uncovered essential mechanisms controlling gene expression in animal cells.
Inositol phosphate metabolites produced by inositol metabolism enzymes serve as vital secondary messengers in eukaryotic cell signaling systems and are broadly implicated in cancer, obesity, diabetes, and neurological disorders.
The research team demonstrated that the inositol polyphosphate multikinase (IPMK) enzyme, a key player in the inositol metabolism system, acts as a critical transcriptional activator within the core gene expression networks of animal cells. Notably, although IPMK was previously reported to play an important role in the transcription process governed by serum response factor (SRF), a representative transcription factor in animal cells, the precise mechanism of its action was unclear.
SRF is a transcription factor directly controlling the expression of at least 200–300 genes, regulating cell growth, proliferation, apoptosis, and motility, and is indispensable for organ development, such as in the heart.
The team discovered that IPMK binds directly to SRF, altering the three-dimensional structure of the SRF protein. This interaction facilitates the transcriptional activity of various genes through the SRF activated by IPMK, demonstrating that IPMK acts as a critical regulatory switch to enhance SRF's protein activity.
< Figure 1. The serum response factor (SRF) protein, a key transcription factor in animal cells, directly binds to inositol polyphosphate multikinase (IPMK) enzyme and undergoes structural change to acquire DNA binding ability, and precisely regulates growth and differentiation of animal cells through transcriptional activation. >
The team further verified that disruptions in the direct interaction between IPMK and SRF lead to the reduced functionality and activity of SRF, causing severe impairments in gene expression.
By highlighting the significance of the intrinsically disordered region (IDR) in SRF, the researchers underscored the biological importance of intrinsically disordered proteins (IDPs). Unlike most proteins that adopt distinct structures through folding, IDPs, including those with IDRs, do not exhibit specific structures but play crucial biological roles, attracting significant attention in the scientific community.
Professor Seyun Kim commented, "This study provides a vital mechanism proving that IPMK, a key enzyme in the inositol metabolism system, is a major transcriptional activator in the core gene expression network of animal cells. By understanding fundamental processes such as cancer development and metastasis, tissue differentiation from stem cells, and neural activation through SRF, we hope this discovery will lead to the broad application of innovative therapeutic technologies."
The findings were published on January 7th in the international journal Nucleic Acids Research (IF=16.7, top 1.8% in Biochemistry and Molecular Biology), under the title “Single-molecule analysis reveals that IPMK enhances the DNA-binding activity of the transcription factor SRF" (DOI: 10.1093/nar/gkae1281).
This research was supported by the National Research Foundation of Korea's Mid-career Research Program, Leading Research Center Program, and Global Research Laboratory Program, as well as by the Suh Kyungbae Science Foundation and the Samsung Future Technology Development Program.
2025.01.24 View 12125
KAIST Uncovers the Principles of Gene Expression Regulation in Cancer and Cellular Functions
< (From left) Professor Seyun Kim, Professor Gwangrog Lee, Dr. Hyoungjoon Ahn, Dr. Jeongmin Yu, Professor Won-Ki Cho, and (below) PhD candidate Kwangmin Ryu of the Department of Biological Sciences>
A research team at KAIST has identified the core gene expression networks regulated by key proteins that fundamentally drive phenomena such as cancer development, metastasis, tissue differentiation from stem cells, and neural activation processes. This discovery lays the foundation for developing innovative therapeutic technologies.
On the 22nd of January, KAIST (represented by President Kwang Hyung Lee) announced that the joint research team led by Professors Seyun Kim, Gwangrog Lee, and Won-Ki Cho from the Department of Biological Sciences had uncovered essential mechanisms controlling gene expression in animal cells.
Inositol phosphate metabolites produced by inositol metabolism enzymes serve as vital secondary messengers in eukaryotic cell signaling systems and are broadly implicated in cancer, obesity, diabetes, and neurological disorders.
The research team demonstrated that the inositol polyphosphate multikinase (IPMK) enzyme, a key player in the inositol metabolism system, acts as a critical transcriptional activator within the core gene expression networks of animal cells. Notably, although IPMK was previously reported to play an important role in the transcription process governed by serum response factor (SRF), a representative transcription factor in animal cells, the precise mechanism of its action was unclear.
SRF is a transcription factor directly controlling the expression of at least 200–300 genes, regulating cell growth, proliferation, apoptosis, and motility, and is indispensable for organ development, such as in the heart.
The team discovered that IPMK binds directly to SRF, altering the three-dimensional structure of the SRF protein. This interaction facilitates the transcriptional activity of various genes through the SRF activated by IPMK, demonstrating that IPMK acts as a critical regulatory switch to enhance SRF's protein activity.
< Figure 1. The serum response factor (SRF) protein, a key transcription factor in animal cells, directly binds to inositol polyphosphate multikinase (IPMK) enzyme and undergoes structural change to acquire DNA binding ability, and precisely regulates growth and differentiation of animal cells through transcriptional activation. >
The team further verified that disruptions in the direct interaction between IPMK and SRF lead to the reduced functionality and activity of SRF, causing severe impairments in gene expression.
By highlighting the significance of the intrinsically disordered region (IDR) in SRF, the researchers underscored the biological importance of intrinsically disordered proteins (IDPs). Unlike most proteins that adopt distinct structures through folding, IDPs, including those with IDRs, do not exhibit specific structures but play crucial biological roles, attracting significant attention in the scientific community.
Professor Seyun Kim commented, "This study provides a vital mechanism proving that IPMK, a key enzyme in the inositol metabolism system, is a major transcriptional activator in the core gene expression network of animal cells. By understanding fundamental processes such as cancer development and metastasis, tissue differentiation from stem cells, and neural activation through SRF, we hope this discovery will lead to the broad application of innovative therapeutic technologies."
The findings were published on January 7th in the international journal Nucleic Acids Research (IF=16.7, top 1.8% in Biochemistry and Molecular Biology), under the title “Single-molecule analysis reveals that IPMK enhances the DNA-binding activity of the transcription factor SRF" (DOI: 10.1093/nar/gkae1281).
This research was supported by the National Research Foundation of Korea's Mid-career Research Program, Leading Research Center Program, and Global Research Laboratory Program, as well as by the Suh Kyungbae Science Foundation and the Samsung Future Technology Development Program.
2025.01.24 View 12125 -
 KAIST Develops CamBio - a New Biotemplating Method
- Professor Jae-Byum Chang and Professor Yeon Sik Jung’s joint research team of the Department of Materials Science and Engineering developed a highly tunable bio-templating method “CamBio” that makes use of intracellular protein structures
- Substrate performance improvement of up to 230% demonstrated via surface-enhanced Raman spectroscopy (SERS)
- Expected to have price competitiveness over bio-templating method as it expands the range of biological samples
- Expected to expand the range of application of nanostructure synthesis technology by utilizing various biological structures
< Photo 1. (From left) Professor Yeon Sik Jung, Ph.D. candidate Dae-Hyeon Song, Professor Jae-Byum Chang, and (from top right) Dr. Chang Woo Song and Dr. Seunghee H. Cho of the Department of Materials Science and Engineering >
Biological structures have complex characteristics that are difficult to replicate artificially, but biotemplating methods* that directly utilize these biological structures have been used in various fields of application. The KAIST research team succeeded in utilizing previously unusable biological structures and expanding the areas in which biotemplate methods can be applied.
*Biotemplating: A method of using biotemplates as a mold to create functional structural materials, utilizing the functions of these biological structures, from viruses to the tissues and organs that make up our bodies
KAIST (President Kwang Hyung Lee) announced on the 10th that a joint research team of Professors Jae-Byum Chang and Professor Yeon Sik Jung of the Department of Materials Science and Engineering developed a biotemplating method that utilizes specific intracellular proteins in biological samples and has high tunability.
Existing biotemplate methods mainly utilize only the external surface of biological samples or have limitations in utilizing the structure-function correlation of various biological structures due to limited dimensions and sample sizes, making it difficult to create functional nanostructures.
To solve this problem, the research team studied a way to utilize various biological structures within the cells while retaining high tunability.
< Figure 1. CamBio utilizing microtubules, a intracellular protein structure. The silver nanoparticle chains synthesized along the microtubules that span the entire cell interior can be observed through an electron microscope, and it is shown that this can be used as a successful SERS substrate. >
As a result of the research, the team developed the “Conversion to advanced materials via labeled Biostructure”, shortened as “CamBio”, which enables the selective synthesis of nanostructures with various characteristics and sizes from specific protein structures composed of diverse proteins within biological specimens.
The CamBio method secures high tunability of functional nanostructures that can be manufactured from biological samples by merging various manufacturing and biological technologies.
Through the technology of repeatedly attaching antibodies, arranging cells in a certain shape, and thinly slicing tissue, the functional nanostructures made with CamBio showed improved performance on the surface-enhanced Raman spectroscopy (SERS)* substrate used for material detection.
*Surface-enhanced Raman spectroscopy (SERS): A technology that can detect very small amounts of substances using light, based on the principle that specific substances react to light and amplifies signals on surfaces of metals such as gold or silver.
The research team found that the nanoparticle chains made using the intracellular protein structures through the process of repeated labeling with antibodies allowed easier control, and improved SERS performance by up to 230%.
In addition, the research team expanded from utilizing the structures inside cells to obtaining samples of muscle tissues inside meat using a cryostat and successfully producing a substrate with periodic bands made of metal particles by performing the CamBio process. This method of producing a substrate not only allows large-scale production using biological samples, but also shows that it is a cost-effective method.
< Figure 2. A method for securing tunability using CamBio at the cell level. Examples of controlling characteristics by integrating iterative labeling and cell pattering techniques with CamBio are shown. >
The CamBio developed by the research team is expected to be used as a way to solve problems faced by various research fields as it is to expand the range of bio-samples that can be produced for various usage.
The first author, Dae-Hyeon Song, a Ph.D. candidate of KAIST Department of Materials Science and Engineering said, “Through CamBio, we have comprehensively accumulated biotemplating methods that can utilize more diverse protein structures,” and “If combined with the state-of-the-art biological technologies such as gene editing and 3D bioprinting and new material synthesis technologies, biostructures can be utilized in various fields of application.”
< Figure 3. A method for securing tunability using CamBio at the tissue level. In order to utilize proteins inside muscle tissue, the frozen tissue sectioning technology is combined, and through this, a substrate with a periodic nanoparticle band pattern is successfully produced, and it is shown that large-area acquisition of samples and price competitiveness can be achieved. >
This study, in which the Ph.D. candidate Dae-Hyeon Song along with Dr. Chang Woo Song, and Dr. Seunghee H. Cho of the same department participated as the first authors, was published online in the international academic journal, Advanced Science, on November 13th, 2024.
(Paper title: Highly Tunable, Nanomaterial-Functionalized Structural Templating of Intracellular Protein Structures Within Biological Species) https://doi.org/10.1002/advs.202406492
This study was conducted with a combination of support from various programs including the National Convergence Research of Scientific Challenges (National Research Foundation of Korea (NRF) 2024), Engineering Reseach Center (ERC) (Wearable Platform Materials Technology Center, NRF 2023), ERC (Global Bio-integrated Materials Center, NRF 2024), and the National Advanced Program for Biological Research Resources (Bioimaging Data Curation Center, NRF 2024) funded by Ministry of Science and ICT.
2025.01.10 View 6591
KAIST Develops CamBio - a New Biotemplating Method
- Professor Jae-Byum Chang and Professor Yeon Sik Jung’s joint research team of the Department of Materials Science and Engineering developed a highly tunable bio-templating method “CamBio” that makes use of intracellular protein structures
- Substrate performance improvement of up to 230% demonstrated via surface-enhanced Raman spectroscopy (SERS)
- Expected to have price competitiveness over bio-templating method as it expands the range of biological samples
- Expected to expand the range of application of nanostructure synthesis technology by utilizing various biological structures
< Photo 1. (From left) Professor Yeon Sik Jung, Ph.D. candidate Dae-Hyeon Song, Professor Jae-Byum Chang, and (from top right) Dr. Chang Woo Song and Dr. Seunghee H. Cho of the Department of Materials Science and Engineering >
Biological structures have complex characteristics that are difficult to replicate artificially, but biotemplating methods* that directly utilize these biological structures have been used in various fields of application. The KAIST research team succeeded in utilizing previously unusable biological structures and expanding the areas in which biotemplate methods can be applied.
*Biotemplating: A method of using biotemplates as a mold to create functional structural materials, utilizing the functions of these biological structures, from viruses to the tissues and organs that make up our bodies
KAIST (President Kwang Hyung Lee) announced on the 10th that a joint research team of Professors Jae-Byum Chang and Professor Yeon Sik Jung of the Department of Materials Science and Engineering developed a biotemplating method that utilizes specific intracellular proteins in biological samples and has high tunability.
Existing biotemplate methods mainly utilize only the external surface of biological samples or have limitations in utilizing the structure-function correlation of various biological structures due to limited dimensions and sample sizes, making it difficult to create functional nanostructures.
To solve this problem, the research team studied a way to utilize various biological structures within the cells while retaining high tunability.
< Figure 1. CamBio utilizing microtubules, a intracellular protein structure. The silver nanoparticle chains synthesized along the microtubules that span the entire cell interior can be observed through an electron microscope, and it is shown that this can be used as a successful SERS substrate. >
As a result of the research, the team developed the “Conversion to advanced materials via labeled Biostructure”, shortened as “CamBio”, which enables the selective synthesis of nanostructures with various characteristics and sizes from specific protein structures composed of diverse proteins within biological specimens.
The CamBio method secures high tunability of functional nanostructures that can be manufactured from biological samples by merging various manufacturing and biological technologies.
Through the technology of repeatedly attaching antibodies, arranging cells in a certain shape, and thinly slicing tissue, the functional nanostructures made with CamBio showed improved performance on the surface-enhanced Raman spectroscopy (SERS)* substrate used for material detection.
*Surface-enhanced Raman spectroscopy (SERS): A technology that can detect very small amounts of substances using light, based on the principle that specific substances react to light and amplifies signals on surfaces of metals such as gold or silver.
The research team found that the nanoparticle chains made using the intracellular protein structures through the process of repeated labeling with antibodies allowed easier control, and improved SERS performance by up to 230%.
In addition, the research team expanded from utilizing the structures inside cells to obtaining samples of muscle tissues inside meat using a cryostat and successfully producing a substrate with periodic bands made of metal particles by performing the CamBio process. This method of producing a substrate not only allows large-scale production using biological samples, but also shows that it is a cost-effective method.
< Figure 2. A method for securing tunability using CamBio at the cell level. Examples of controlling characteristics by integrating iterative labeling and cell pattering techniques with CamBio are shown. >
The CamBio developed by the research team is expected to be used as a way to solve problems faced by various research fields as it is to expand the range of bio-samples that can be produced for various usage.
The first author, Dae-Hyeon Song, a Ph.D. candidate of KAIST Department of Materials Science and Engineering said, “Through CamBio, we have comprehensively accumulated biotemplating methods that can utilize more diverse protein structures,” and “If combined with the state-of-the-art biological technologies such as gene editing and 3D bioprinting and new material synthesis technologies, biostructures can be utilized in various fields of application.”
< Figure 3. A method for securing tunability using CamBio at the tissue level. In order to utilize proteins inside muscle tissue, the frozen tissue sectioning technology is combined, and through this, a substrate with a periodic nanoparticle band pattern is successfully produced, and it is shown that large-area acquisition of samples and price competitiveness can be achieved. >
This study, in which the Ph.D. candidate Dae-Hyeon Song along with Dr. Chang Woo Song, and Dr. Seunghee H. Cho of the same department participated as the first authors, was published online in the international academic journal, Advanced Science, on November 13th, 2024.
(Paper title: Highly Tunable, Nanomaterial-Functionalized Structural Templating of Intracellular Protein Structures Within Biological Species) https://doi.org/10.1002/advs.202406492
This study was conducted with a combination of support from various programs including the National Convergence Research of Scientific Challenges (National Research Foundation of Korea (NRF) 2024), Engineering Reseach Center (ERC) (Wearable Platform Materials Technology Center, NRF 2023), ERC (Global Bio-integrated Materials Center, NRF 2024), and the National Advanced Program for Biological Research Resources (Bioimaging Data Curation Center, NRF 2024) funded by Ministry of Science and ICT.
2025.01.10 View 6591 -
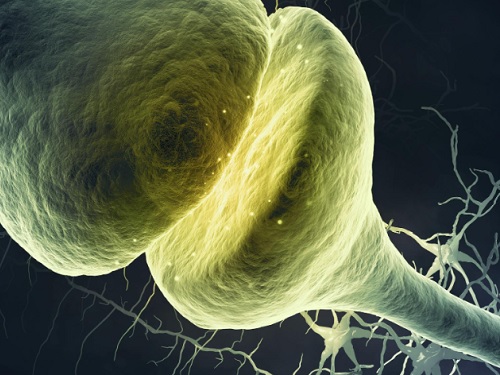 A KAIST Research Team Observes the Processes of Memory and Cognition in Real Time
The human brain contains approximately 86 billion neurons and 600 trillion synapses that exchange signals between the neurons to help us control the various functions of the brain including cognition, emotion, and memory. Interestingly, the number of synapses decrease with age or as a result of diseases like Alzheimer’s, and research on synapses thus attracts a lot of attention. However, limitations have existed in observing the dynamics of synapse structures in real time.
On January 9, a joint research team led by Professor Won Do Heo from the KAIST Department of Biological Sciences, Professor Hyung-Bae Kwon from Johns Hopkins School of Medicine, and Professor Sangkyu Lee from the Institute for Basic Science (IBS) revealed that they have developed the world’s first technique to allow a real-time observation of synapse formation, extinction, and alterations.
Professor Heo’s team conjugated dimerization-dependent fluorescent proteins (ddFP) to synapses in order to observe the process in which synapses create connections between neurons in real time. The team named this technique SynapShot, by combining the words ‘synapse’ and snapshot’, and successfully tracked and observed the live formation and extinction processes of synapses as well as their dynamic changes.
< Figure 1. To observe dynamically changing synapses, dimerization-dependent fluorescent protein (ddFP) was expressed to observe flourescent signals upon synapse formation as ddFP enables fluorescence detection through reversible binding to pre- and postsynaptic terminals. >
Through a joint research project, the teams led by Professor Heo and Professor Sangkyu Lee at IBS together designed a SynapShot with green and red fluorescence, and were able to easily distinguish the synapse connecting two different neurons. Additionally, by combining an optogenetic technique that can control the function of a molecule using light, the team was able to observe the changes in the synapses while simultaneously inducing certain functions of the neurons using light.
Through more joint research with the team led by Professor Hyung-Bae Kwon at the Johns Hopkins School of Medicine, Professor Heo’s team induced several situations on live mice, including visual discrimination training, exercise, and anaesthesia, and used SynapShot to observe the changes in the synapses during each situation in real time. The observations revealed that each synapse could change fairly quickly and dynamically. This was the first-ever case in which the changes in synapses were observed in a live mammal.
< Figure 2. Microscopic photos observed through changes of the flourescence of the synapse sensor (SynapShot) by cultivating the neurons of an experimental rat and expressing the SynapShot. The changes in the synapse that is created when the pre- and post-synaptic terminals come into contact and the synapse that disappears after a certain period of time are measured by the fluorescence of the SynapShot. >
Professor Heo said, “Our group developed SynapShot through a collaboration with domestic and international research teams, and have opened up the possibility for first-hand live observations of the quick and dynamic changes of synapses, which was previously difficult to do. We expect this technique to revolutionize research methodology in the neurological field, and play an important role in brightening the future of brain science.”
This research, conducted by co-first authors Seungkyu Son (Ph.D. candidate), Jinsu Lee (Ph.D. candidate) and Dr. Kanghoon Jung from Johns Hopkins, was published in the online edition of Nature Methods on January 8 under the title “Real-time visualization of structural dynamics of synapses in live cells in vivo”, and will be printed in the February volume.
< Figure 3. Simultaneous use of green-SynapShot and red-SynapShot to distinguish and observe synapses with one post-terminal and different pre-terminals. >
< Figure 4. Dimer-dependent fluorescent protein (ddFP) exists as a green fluorescent protein as well as a red fluorescent protein, and can be applied together with blue light-activated optogenetic technology. After activating Tropomyosin receptor kinase B (TrkB) by blue light using optogenetic technology, the strengthening of synaptic connections through signals of brain-derived neurotrophic factor is observed using red-SynapShot. >
< Figure 5. Micrographs showing real-time changing synapses in the visual cortex of mice trained through visual training using in vivo imaging techniques such as two-photon microscopy as well as at the cellular level. >
This research was supported by Mid-Sized Research Funds and the Singularity Project from KAIST, and by IBS.
2024.01.18 View 8561
A KAIST Research Team Observes the Processes of Memory and Cognition in Real Time
The human brain contains approximately 86 billion neurons and 600 trillion synapses that exchange signals between the neurons to help us control the various functions of the brain including cognition, emotion, and memory. Interestingly, the number of synapses decrease with age or as a result of diseases like Alzheimer’s, and research on synapses thus attracts a lot of attention. However, limitations have existed in observing the dynamics of synapse structures in real time.
On January 9, a joint research team led by Professor Won Do Heo from the KAIST Department of Biological Sciences, Professor Hyung-Bae Kwon from Johns Hopkins School of Medicine, and Professor Sangkyu Lee from the Institute for Basic Science (IBS) revealed that they have developed the world’s first technique to allow a real-time observation of synapse formation, extinction, and alterations.
Professor Heo’s team conjugated dimerization-dependent fluorescent proteins (ddFP) to synapses in order to observe the process in which synapses create connections between neurons in real time. The team named this technique SynapShot, by combining the words ‘synapse’ and snapshot’, and successfully tracked and observed the live formation and extinction processes of synapses as well as their dynamic changes.
< Figure 1. To observe dynamically changing synapses, dimerization-dependent fluorescent protein (ddFP) was expressed to observe flourescent signals upon synapse formation as ddFP enables fluorescence detection through reversible binding to pre- and postsynaptic terminals. >
Through a joint research project, the teams led by Professor Heo and Professor Sangkyu Lee at IBS together designed a SynapShot with green and red fluorescence, and were able to easily distinguish the synapse connecting two different neurons. Additionally, by combining an optogenetic technique that can control the function of a molecule using light, the team was able to observe the changes in the synapses while simultaneously inducing certain functions of the neurons using light.
Through more joint research with the team led by Professor Hyung-Bae Kwon at the Johns Hopkins School of Medicine, Professor Heo’s team induced several situations on live mice, including visual discrimination training, exercise, and anaesthesia, and used SynapShot to observe the changes in the synapses during each situation in real time. The observations revealed that each synapse could change fairly quickly and dynamically. This was the first-ever case in which the changes in synapses were observed in a live mammal.
< Figure 2. Microscopic photos observed through changes of the flourescence of the synapse sensor (SynapShot) by cultivating the neurons of an experimental rat and expressing the SynapShot. The changes in the synapse that is created when the pre- and post-synaptic terminals come into contact and the synapse that disappears after a certain period of time are measured by the fluorescence of the SynapShot. >
Professor Heo said, “Our group developed SynapShot through a collaboration with domestic and international research teams, and have opened up the possibility for first-hand live observations of the quick and dynamic changes of synapses, which was previously difficult to do. We expect this technique to revolutionize research methodology in the neurological field, and play an important role in brightening the future of brain science.”
This research, conducted by co-first authors Seungkyu Son (Ph.D. candidate), Jinsu Lee (Ph.D. candidate) and Dr. Kanghoon Jung from Johns Hopkins, was published in the online edition of Nature Methods on January 8 under the title “Real-time visualization of structural dynamics of synapses in live cells in vivo”, and will be printed in the February volume.
< Figure 3. Simultaneous use of green-SynapShot and red-SynapShot to distinguish and observe synapses with one post-terminal and different pre-terminals. >
< Figure 4. Dimer-dependent fluorescent protein (ddFP) exists as a green fluorescent protein as well as a red fluorescent protein, and can be applied together with blue light-activated optogenetic technology. After activating Tropomyosin receptor kinase B (TrkB) by blue light using optogenetic technology, the strengthening of synaptic connections through signals of brain-derived neurotrophic factor is observed using red-SynapShot. >
< Figure 5. Micrographs showing real-time changing synapses in the visual cortex of mice trained through visual training using in vivo imaging techniques such as two-photon microscopy as well as at the cellular level. >
This research was supported by Mid-Sized Research Funds and the Singularity Project from KAIST, and by IBS.
2024.01.18 View 8561 -
 KAIST research team develops clathrin assembly for targeted protein delivery to cancer cells
In order to effectively treat cancer without additional side effects, we need a way to deliver drugs specifically to tumor cells. Protein assemblies have been widely used for drug delivery in the field of cancer treatment, but to use them for drug delivery they must first be functionalized, meaning they must be bound to the protein that recognizes the target tumor cell and deliver a drug that kills it. However, the functionalization process of protein assemblies is very complex, inefficient, and limited to small-sized chemical drugs, which limits their real-life applicability.
On March 14, a KAIST research team led by Professor Hak-Sung Kim from the KAIST Department of Biological Sciences reported the development of a clathrin assembly that can specifically deliver drugs to cancer cells.
Clathrin assemblies transport materials efficiently through endocytosis in living organisms. They are formed by the self-assembly of triskelion units, which are composed of three heavy chains bonded with three light chains. Inspired by this mechanism, the research team designed a clathrin chain to facilitate the functionalization of tumor cell recognition proteins and toxin proteins in order to deliver drugs specifically to tumor cells. From this, the team created a new type of clathrin assembly.
Figure 1. (Upper) Schematic diagram of the development of a new clathrin assembly that simultaneously functionalizes two types of proteins (cancer cell recognition protein and toxin protein) on heavy and light chains of clathrin in a one-pot reaction (bottom, left) Electron microscopy image of clathrin assembly: formation of an assembly with a diameter of about 28 nanometers (bottom, right) Cancer cell killing effect of CLA: CLA functionalized with epidermal growth factor receptor (EGFR) recognition protein and toxin protein kills only the cancer cells that overexpress EGFR.
The newly developed clathrin assembly requires a one-pot reaction, meaning both the toxin and tumor-recognition proteins can be functionalized simultaneously and show high efficiency. As a result, this technique is expected to be used in a wide variety of applications in the fields of biology and medicine including drug delivery, vaccine development, and diagnosing illnesses.
In this research, an epidermal growth factor receptor (EGFR), a common tumor marker, was used as the recognition protein, allowing drug delivery only to tumor cells. The clathrin assemblies that were functionalized to recognize EGFR showed a bonding strength 900-times stronger than it normally would due to the avidity effect. Based on this finding, the research team confirmed that treatment with toxin-functionalized clathrin assembly led to effective cell death for tumor cells, while it showed no such effect on healthy cells.
This research by Dr. Hong-Sik Kim and his colleagues was published in Small volume 19, issue 8 on February 22 under the title, "Construction and Functionalization of a Clathrin Assembly for a Targeted Protein Delivery", and it was selected as the cover paper.
Figure 2. Cover Paper: This study was published in the international journal 'Small' on February 22nd, Volume 19, No. 8, and was selected as the cover paper.
First author Dr. Hong-Sik Kim said, “Clathrin is difficult to functionalize, and since it is extracted from mammals, realistic applications have been limited.” He added, “But the new clathrin assembly we designed for this research can be functionalized with two different types of proteins through a single-step reaction, and can be produced from E. coli, meaning it can become an applicable protein assembly technology for a wide range of biomedical fields.”
This research was funded by the Global Ph.D. Fellowship and the Mid-career Researcher Grant of the National Research Foundation.
2023.03.22 View 8413
KAIST research team develops clathrin assembly for targeted protein delivery to cancer cells
In order to effectively treat cancer without additional side effects, we need a way to deliver drugs specifically to tumor cells. Protein assemblies have been widely used for drug delivery in the field of cancer treatment, but to use them for drug delivery they must first be functionalized, meaning they must be bound to the protein that recognizes the target tumor cell and deliver a drug that kills it. However, the functionalization process of protein assemblies is very complex, inefficient, and limited to small-sized chemical drugs, which limits their real-life applicability.
On March 14, a KAIST research team led by Professor Hak-Sung Kim from the KAIST Department of Biological Sciences reported the development of a clathrin assembly that can specifically deliver drugs to cancer cells.
Clathrin assemblies transport materials efficiently through endocytosis in living organisms. They are formed by the self-assembly of triskelion units, which are composed of three heavy chains bonded with three light chains. Inspired by this mechanism, the research team designed a clathrin chain to facilitate the functionalization of tumor cell recognition proteins and toxin proteins in order to deliver drugs specifically to tumor cells. From this, the team created a new type of clathrin assembly.
Figure 1. (Upper) Schematic diagram of the development of a new clathrin assembly that simultaneously functionalizes two types of proteins (cancer cell recognition protein and toxin protein) on heavy and light chains of clathrin in a one-pot reaction (bottom, left) Electron microscopy image of clathrin assembly: formation of an assembly with a diameter of about 28 nanometers (bottom, right) Cancer cell killing effect of CLA: CLA functionalized with epidermal growth factor receptor (EGFR) recognition protein and toxin protein kills only the cancer cells that overexpress EGFR.
The newly developed clathrin assembly requires a one-pot reaction, meaning both the toxin and tumor-recognition proteins can be functionalized simultaneously and show high efficiency. As a result, this technique is expected to be used in a wide variety of applications in the fields of biology and medicine including drug delivery, vaccine development, and diagnosing illnesses.
In this research, an epidermal growth factor receptor (EGFR), a common tumor marker, was used as the recognition protein, allowing drug delivery only to tumor cells. The clathrin assemblies that were functionalized to recognize EGFR showed a bonding strength 900-times stronger than it normally would due to the avidity effect. Based on this finding, the research team confirmed that treatment with toxin-functionalized clathrin assembly led to effective cell death for tumor cells, while it showed no such effect on healthy cells.
This research by Dr. Hong-Sik Kim and his colleagues was published in Small volume 19, issue 8 on February 22 under the title, "Construction and Functionalization of a Clathrin Assembly for a Targeted Protein Delivery", and it was selected as the cover paper.
Figure 2. Cover Paper: This study was published in the international journal 'Small' on February 22nd, Volume 19, No. 8, and was selected as the cover paper.
First author Dr. Hong-Sik Kim said, “Clathrin is difficult to functionalize, and since it is extracted from mammals, realistic applications have been limited.” He added, “But the new clathrin assembly we designed for this research can be functionalized with two different types of proteins through a single-step reaction, and can be produced from E. coli, meaning it can become an applicable protein assembly technology for a wide range of biomedical fields.”
This research was funded by the Global Ph.D. Fellowship and the Mid-career Researcher Grant of the National Research Foundation.
2023.03.22 View 8413 -
 A Genetic Change for Achieving a Long and Healthy Life
Researchers identified a single amino acid change in the tumor suppressor protein in PTEN that extends healthy periods while maintaining longevity
Living a long, healthy life is everyone’s wish, but it is not an easy one to achieve. Many aging studies are developing strategies to increase health spans, the period of life spent with good health, without chronic diseases and disabilities. Researchers at KAIST presented new insights for improving the health span by just regulating the activity of a protein.
A research group under Professor Seung-Jae V. Lee from the Department of Biological Sciences identified a single amino acid change in the tumor suppressor protein phosphatase and tensin homolog (PTEN) that dramatically extends healthy periods while maintaining longevity. This study highlights the importance of the well-conserved tumor suppressor protein PTEN in health span regulation, which can be targeted to develop therapies for promoting healthy longevity in humans. The research was published in Nature Communications on September 24, 2021.
Insulin and insulin-like growth factor-1 (IGF-1) signaling (IIS) is one of the evolutionarily conserved aging-modulatory pathways present in life forms ranging from tiny roundworms to humans. The proper reduction of IIS leads to longevity in animals but often causes defects in multiple health parameters including impaired motility, reproduction, and growth.
The research team found that a specific amino acid change in the PTEN protein improves health status while retaining the longevity conferred by reduced IIS. They used the roundworm C. elegans, an excellent model animal that has been widely used for aging research, mainly because of its very short normal lifespan of about two to three weeks. The PTEN protein is a phosphatase that removes phosphate from lipids as well as proteins. Interestingly, the newly identified amino acid change delicately recalibrated the IIS by partially maintaining protein phosphatase activity while reducing lipid phosphatase activity.
As a result, the amino acid change in the PTEN protein maintained the activity of the longevity-promoting transcription factor Forkhead Box O (FOXO) protein while restricting the detrimental upregulation of another transcription factor, NRF2, leading to long and healthy life in animals with reduced IIS.
Professor Lee said, “Our study raises the exciting possibility of simultaneously promoting longevity and health in humans by slightly tweaking the activity of one protein, PTEN.”
This work was supported by the MInistry of Science and ICT through the National Research Foundation of Korea.
-Publication:Hae-Eun H. Park, Wooseon Hwang, Seokjin Ham, Eunah Kim, Ozlem Altintas, Sangsoon Park, Heehwa G. Son, Yujin Lee, Dongyeop Lee, Won Do Heo, and Seung-Jae V. Lee. 2021. “A PTEN variant uncouples longevity from impaired fitness in Caenorhabditis elegans with reduced insulin/IGF-1 signaling,” Nature Communications, 12(1), 5631. (https://doi.org/10.1038/s41467-021-25920-w)
-ProfileProfessor Seung-Jae V. LeeMolecular Genetics of Aging LaboratoryDepartment of Biological Sciences KAIST
2021.11.19 View 10413
A Genetic Change for Achieving a Long and Healthy Life
Researchers identified a single amino acid change in the tumor suppressor protein in PTEN that extends healthy periods while maintaining longevity
Living a long, healthy life is everyone’s wish, but it is not an easy one to achieve. Many aging studies are developing strategies to increase health spans, the period of life spent with good health, without chronic diseases and disabilities. Researchers at KAIST presented new insights for improving the health span by just regulating the activity of a protein.
A research group under Professor Seung-Jae V. Lee from the Department of Biological Sciences identified a single amino acid change in the tumor suppressor protein phosphatase and tensin homolog (PTEN) that dramatically extends healthy periods while maintaining longevity. This study highlights the importance of the well-conserved tumor suppressor protein PTEN in health span regulation, which can be targeted to develop therapies for promoting healthy longevity in humans. The research was published in Nature Communications on September 24, 2021.
Insulin and insulin-like growth factor-1 (IGF-1) signaling (IIS) is one of the evolutionarily conserved aging-modulatory pathways present in life forms ranging from tiny roundworms to humans. The proper reduction of IIS leads to longevity in animals but often causes defects in multiple health parameters including impaired motility, reproduction, and growth.
The research team found that a specific amino acid change in the PTEN protein improves health status while retaining the longevity conferred by reduced IIS. They used the roundworm C. elegans, an excellent model animal that has been widely used for aging research, mainly because of its very short normal lifespan of about two to three weeks. The PTEN protein is a phosphatase that removes phosphate from lipids as well as proteins. Interestingly, the newly identified amino acid change delicately recalibrated the IIS by partially maintaining protein phosphatase activity while reducing lipid phosphatase activity.
As a result, the amino acid change in the PTEN protein maintained the activity of the longevity-promoting transcription factor Forkhead Box O (FOXO) protein while restricting the detrimental upregulation of another transcription factor, NRF2, leading to long and healthy life in animals with reduced IIS.
Professor Lee said, “Our study raises the exciting possibility of simultaneously promoting longevity and health in humans by slightly tweaking the activity of one protein, PTEN.”
This work was supported by the MInistry of Science and ICT through the National Research Foundation of Korea.
-Publication:Hae-Eun H. Park, Wooseon Hwang, Seokjin Ham, Eunah Kim, Ozlem Altintas, Sangsoon Park, Heehwa G. Son, Yujin Lee, Dongyeop Lee, Won Do Heo, and Seung-Jae V. Lee. 2021. “A PTEN variant uncouples longevity from impaired fitness in Caenorhabditis elegans with reduced insulin/IGF-1 signaling,” Nature Communications, 12(1), 5631. (https://doi.org/10.1038/s41467-021-25920-w)
-ProfileProfessor Seung-Jae V. LeeMolecular Genetics of Aging LaboratoryDepartment of Biological Sciences KAIST
2021.11.19 View 10413 -
 The Dynamic Tracking of Tissue-Specific Secretory Proteins
Researchers develop a versatile and powerful tool for studying the spatiotemporal dynamics of secretory proteins, a valuable class of biomarkers and therapeutic targets
Researchers have presented a method for profiling tissue-specific secretory proteins in live mice. This method is expected to be applicable to various tissues or disease models for investigating biomarkers or therapeutic targets involved in disease progression. This research was reported in Nature Communications on September 1.
Secretory proteins released into the blood play essential roles in physiological systems. They are core mediators of interorgan communication, while serving as biomarkers and therapeutic targets.
Previous studies have analyzed conditioned media from culture models to identify cell type-specific secretory proteins, but these models often fail to fully recapitulate the intricacies of multi-organ systems and thus do not sufficiently reflect biological realities.
These limitations provided compelling motivation for the research team led by Jae Myoung Suh and his collaborators to develop techniques that could identify and resolve characteristics of tissue-specific secretory proteins along time and space dimensions.
For addressing this gap in the current methodology, the research team utilized proximity-labeling enzymes such as TurboID to label secretory proteins in endoplasmic reticulum lumen using biotin. Thereafter, the biotin-labeled secretory proteins were readily enriched through streptavidin affinity purification and could be identified through mass spectrometry.
To demonstrate its functionality in live mice, research team delivered TurboID to mouse livers via an adenovirus. After administering the biotin, only liver-derived secretory proteins were successfully detected in the plasma of the mice. Interestingly, the pattern of biotin-labeled proteins secreted from the liver was clearly distinctive from those of hepatocyte cell lines.
First author Kwang-eun Kim from the Graduate School of Medical Science and Engineering explained, “The proteins secreted by the liver were significantly different from the results of cell culture models. This data shows the limitations of cell culture models for secretory protein study, and this technique can overcome those limitations. It can be further used to discover biomarkers and therapeutic targets that can more fully reflect the physiological state.”
This work research was supported by the National Research Foundation of Korea, the KAIST Key Research Institutes Project (Interdisciplinary Research Group), and the Institute for Basic Science in Korea.
-PublicationKwang-eun Kim, Isaac Park et al., “Dynamic tracking and identification of tissue-specific secretory proteins in the circulation of live mice,” Nature Communications on Sept.1,
2021(https://doi.org/10.1038/s41467-021-25546-y)
-ProfileProfessor Jae Myoung Suh Integrated Lab of Metabolism, Obesity and Diabetes Researchhttps://imodkaist.wixsite.com/home
Graduate School of Medical Science and Engineering College of Life Science and BioengineeringKAIST
2021.09.14 View 11357
The Dynamic Tracking of Tissue-Specific Secretory Proteins
Researchers develop a versatile and powerful tool for studying the spatiotemporal dynamics of secretory proteins, a valuable class of biomarkers and therapeutic targets
Researchers have presented a method for profiling tissue-specific secretory proteins in live mice. This method is expected to be applicable to various tissues or disease models for investigating biomarkers or therapeutic targets involved in disease progression. This research was reported in Nature Communications on September 1.
Secretory proteins released into the blood play essential roles in physiological systems. They are core mediators of interorgan communication, while serving as biomarkers and therapeutic targets.
Previous studies have analyzed conditioned media from culture models to identify cell type-specific secretory proteins, but these models often fail to fully recapitulate the intricacies of multi-organ systems and thus do not sufficiently reflect biological realities.
These limitations provided compelling motivation for the research team led by Jae Myoung Suh and his collaborators to develop techniques that could identify and resolve characteristics of tissue-specific secretory proteins along time and space dimensions.
For addressing this gap in the current methodology, the research team utilized proximity-labeling enzymes such as TurboID to label secretory proteins in endoplasmic reticulum lumen using biotin. Thereafter, the biotin-labeled secretory proteins were readily enriched through streptavidin affinity purification and could be identified through mass spectrometry.
To demonstrate its functionality in live mice, research team delivered TurboID to mouse livers via an adenovirus. After administering the biotin, only liver-derived secretory proteins were successfully detected in the plasma of the mice. Interestingly, the pattern of biotin-labeled proteins secreted from the liver was clearly distinctive from those of hepatocyte cell lines.
First author Kwang-eun Kim from the Graduate School of Medical Science and Engineering explained, “The proteins secreted by the liver were significantly different from the results of cell culture models. This data shows the limitations of cell culture models for secretory protein study, and this technique can overcome those limitations. It can be further used to discover biomarkers and therapeutic targets that can more fully reflect the physiological state.”
This work research was supported by the National Research Foundation of Korea, the KAIST Key Research Institutes Project (Interdisciplinary Research Group), and the Institute for Basic Science in Korea.
-PublicationKwang-eun Kim, Isaac Park et al., “Dynamic tracking and identification of tissue-specific secretory proteins in the circulation of live mice,” Nature Communications on Sept.1,
2021(https://doi.org/10.1038/s41467-021-25546-y)
-ProfileProfessor Jae Myoung Suh Integrated Lab of Metabolism, Obesity and Diabetes Researchhttps://imodkaist.wixsite.com/home
Graduate School of Medical Science and Engineering College of Life Science and BioengineeringKAIST
2021.09.14 View 11357 -
 X-ray Scattering Shines Light on Protein Folding
- Multiple forms of a non-functional, unfolded protein follow different pathways and timelines to reach its folded, functional state, a study reveals. -
KAIST researchers have used an X-ray method to track how proteins fold, which could improve computer simulations of this process, with implications for understanding diseases and improving drug discovery. Their findings were reported in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 30.
When proteins are translated from their DNA codes, they quickly transform from a non-functional, unfolded state into their folded, functional state. Problems in folding can lead to diseases like Alzheimer’s and Parkinson’s.
“Protein folding is one of the most important biological processes, as it forms the functioning 3D protein structure,” explained the physical chemist Hyotcherl Ihee of the Department of Chemistry at KAIST. Dr. Tae Wu Kim, the lead author of this research from Ihee’s group, added, “Understanding the mechanisms of protein folding is important, and could pave the way for disease study and drug development.”
Ihee’s team developed an approach using an X-ray scattering technique to uncover how the protein cytochrome c folds from its initial unfolded state. This protein is composed of a chain of 104 amino acids with an iron-containing heme molecule. It is often used for protein folding studies.
The researchers placed the protein in a solution and shined ultraviolet light on it. This process provides electrons to cytochrome c, reducing the iron within it from the ferric to the ferrous form, which initiates folding. As this was happening, the researchers beamed X-rays at very short intervals onto the sample. The X-rays scattered off all the atomic pairs in the sample and a detector continuously recorded the X-ray scattering patterns. The X-ray scattering patterns provided direct information regarding the 3D protein structure and the changes made in these patterns over time showed real-time motion of the protein during the folding process.
The team found cytochrome c proteins initially exist in a wide variety of unfolded states. Once the folding process is triggered, they stop by a group of intermediates within 31.6 microseconds, and then those intermediates follow different pathways with different folding times to reach an energetically stable folded state.
“We don’t know if this diversity in folding paths can be generalized to other proteins,” Ihee confessed. He continued, “However, we believe that our approach can be used to study other protein folding systems.”
Ihee hopes this approach can improve the accuracy of models that simulate protein interactions by including information on their unstructured states. These simulations are important as they can help identify barriers to proper folding and predict a protein’s folded state given its amino acid sequence. Ultimately, the models could help clarify how some diseases develop and how drugs interact with various protein structures.
Ihee’s group collaborated with Professor Young Min Rhee at the KAIST Department of Chemistry, and this work was supported by the National Research Foundation of Korea (NRF) and the Institute for Basic Science (IBS).
Figure. The scientists found that non-functional unfolded forms of the protein cytochrome c follow different pathways and timelines to reach a stable functional folded state.
Publications:
Kim, T. W., et al. (2020) ‘Protein folding from heterogeneous unfolded state revealed by time-resolved X-ray solution scattering’. PNAS. Volume 117. Issue 26. Page 14996-15005. Available online at https://doi.org/10.1073/pnas.1913442117
Profile: Hyotcherl Ihee, Ph.D.
Professor
hyotcherl.ihee@kaist.ac.kr
http://time.kaist.ac.kr/
Ihee Laboratory
Department of Chemistry
KAIST
https://www.kaist.ac.kr
Daejeon 34141, Korea
Profile: Young Min Rhee, Ph.D.
Professor
ymrhee@kaist.ac.kr
http://singlet.kaist.ac.kr
Rhee Research Group
Department of Chemistry
KAIST
https://www.kaist.ac.kr
Daejeon 34141, Korea
(END)
2020.07.09 View 18185
X-ray Scattering Shines Light on Protein Folding
- Multiple forms of a non-functional, unfolded protein follow different pathways and timelines to reach its folded, functional state, a study reveals. -
KAIST researchers have used an X-ray method to track how proteins fold, which could improve computer simulations of this process, with implications for understanding diseases and improving drug discovery. Their findings were reported in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 30.
When proteins are translated from their DNA codes, they quickly transform from a non-functional, unfolded state into their folded, functional state. Problems in folding can lead to diseases like Alzheimer’s and Parkinson’s.
“Protein folding is one of the most important biological processes, as it forms the functioning 3D protein structure,” explained the physical chemist Hyotcherl Ihee of the Department of Chemistry at KAIST. Dr. Tae Wu Kim, the lead author of this research from Ihee’s group, added, “Understanding the mechanisms of protein folding is important, and could pave the way for disease study and drug development.”
Ihee’s team developed an approach using an X-ray scattering technique to uncover how the protein cytochrome c folds from its initial unfolded state. This protein is composed of a chain of 104 amino acids with an iron-containing heme molecule. It is often used for protein folding studies.
The researchers placed the protein in a solution and shined ultraviolet light on it. This process provides electrons to cytochrome c, reducing the iron within it from the ferric to the ferrous form, which initiates folding. As this was happening, the researchers beamed X-rays at very short intervals onto the sample. The X-rays scattered off all the atomic pairs in the sample and a detector continuously recorded the X-ray scattering patterns. The X-ray scattering patterns provided direct information regarding the 3D protein structure and the changes made in these patterns over time showed real-time motion of the protein during the folding process.
The team found cytochrome c proteins initially exist in a wide variety of unfolded states. Once the folding process is triggered, they stop by a group of intermediates within 31.6 microseconds, and then those intermediates follow different pathways with different folding times to reach an energetically stable folded state.
“We don’t know if this diversity in folding paths can be generalized to other proteins,” Ihee confessed. He continued, “However, we believe that our approach can be used to study other protein folding systems.”
Ihee hopes this approach can improve the accuracy of models that simulate protein interactions by including information on their unstructured states. These simulations are important as they can help identify barriers to proper folding and predict a protein’s folded state given its amino acid sequence. Ultimately, the models could help clarify how some diseases develop and how drugs interact with various protein structures.
Ihee’s group collaborated with Professor Young Min Rhee at the KAIST Department of Chemistry, and this work was supported by the National Research Foundation of Korea (NRF) and the Institute for Basic Science (IBS).
Figure. The scientists found that non-functional unfolded forms of the protein cytochrome c follow different pathways and timelines to reach a stable functional folded state.
Publications:
Kim, T. W., et al. (2020) ‘Protein folding from heterogeneous unfolded state revealed by time-resolved X-ray solution scattering’. PNAS. Volume 117. Issue 26. Page 14996-15005. Available online at https://doi.org/10.1073/pnas.1913442117
Profile: Hyotcherl Ihee, Ph.D.
Professor
hyotcherl.ihee@kaist.ac.kr
http://time.kaist.ac.kr/
Ihee Laboratory
Department of Chemistry
KAIST
https://www.kaist.ac.kr
Daejeon 34141, Korea
Profile: Young Min Rhee, Ph.D.
Professor
ymrhee@kaist.ac.kr
http://singlet.kaist.ac.kr
Rhee Research Group
Department of Chemistry
KAIST
https://www.kaist.ac.kr
Daejeon 34141, Korea
(END)
2020.07.09 View 18185