Department+of+Mathematical+Sciences
-
 Mathematical Modeling Makes a Breakthrough for a New CRSD Medication
PhD Candidate Dae Wook Kim (Left) and Professor Jae Kyoung Kim (Right)
- Systems approach reveals photosensitivity and PER2 level as determinants of clock-modulator efficacy -
Mathematicians’ new modeling has identified major sources of interspecies and inter-individual variations in the clinical efficacy of a clock-modulating drug: photosensitivity and PER2 level. This enabled precision medicine for circadian disruption.
A KAIST mathematics research team led by Professor Jae Kyoung Kim, in collaboration with Pfizer, applied a combination of mathematical modeling and simulation tools for circadian rhythms sleep disorders (CRSDs) to analyze the animal data generated by Pfizer. This study was reported in Molecular Systems Biology as the cover article on July 8.
Pharmaceutical companies have conducted extensive studies on animals to determine the candidacy of this new medication. However, the results of animal testing do not always translate to the same effects in human trials. Furthermore, even between humans, efficacy differs across individuals depending on an individual’s genetic and environmental factors, which require different treatment strategies.
To overcome these obstacles, KAIST mathematicians and their collaborators developed adaptive chronotherapeutics to identify precise dosing regimens that could restore normal circadian phase under different conditions.
A circadian rhythm is a 24-hour cycle in the physiological processes of living creatures, including humans. A biological clock in the hypothalamic suprachiasmatic nucleus in the human brain sets the time for various human behaviors such as sleep.
A disruption of the endogenous timekeeping system caused by changes in one’s life pattern leads to advanced or delayed sleep-wake cycle phase and a desynchronization between sleep-wake rhythms, resulting in CRSDs. To restore the normal timing of sleep, timing of the circadian clock could be adjusted pharmacologically.
Pfizer identified PF-670462, which can adjust the timing of circadian clock by inhibiting the core clock kinase of the circadian clock (CK1d/e). However, the efficacy of PF-670462 significantly differs between nocturnal mice and diurnal monkeys, whose sleeping times are opposite.
The research team discovered the source of such interspecies variations in drug response by performing thousands of virtual experiments using a mathematical model, which describes biochemical interactions among clock molecules and PF-670462. The result suggests that the effect of PF-670462 is reduced by light exposure in diurnal primates more than in nocturnal mice. This indicates that the strong counteracting effect of light must be considered in order to effectively regulate the circadian clock of diurnal humans using PF-670462.
Furthermore, the team also found the source of inter-patients variations in drug efficacy using virtual patients whose circadian clocks were disrupted due to various mutations. The degree of perturbation in the endogenous level of the core clock molecule PER2 affects the efficacy.
This explains why the clinical outcomes of clock-modulating drugs are highly variable and certain subtypes are unresponsive to treatment. Furthermore, this points out the limitations of current treatment strategies tailored to only the patient’s sleep and wake time but not to the molecular cause of sleep disorders.
PhD candidate Dae Wook Kim, who is the first author, said that this motivates the team to develop an adaptive chronotherapy, which identifies a personalized optimal dosing time of day by tracking the sleep-wake up time of patients via a wearable device and allows for a precision medicine approach for CRSDs.
Professor Jae Kyoung Kim said, "As a mathematician, I am excited to help enable the advancement of a new drug candidate, which can improve the lives of so many patients. I hope this result promotes more collaborations in this translational research.”
This research was supported by a Pfizer grant to KAIST (G01160179), the Human Frontiers Science Program Organization (RGY0063/2017), and a National Research Foundation (NRF) of Korea Grant (NRF-2016 RICIB 3008468 and NRF-2017-Fostering Core Leaders of the Future Basic Science Program/ Global Ph.D. Fellowship Program).
Figure 1. Interspecies and Inter-patients Variations in PF-670462 Efficacy
Figure 2. Journal Cover Page
Publication:
Dae Wook Kim, Cheng Chang, Xian Chen, Angela C Doran, Francois Gaudreault, Travis Wager, George J DeMarco, and Jae Kyoung Kim. 2019. Systems approach reveals photosensitivity and PER2 level as determinants of clock-modulator efficacy. Molecular Systems Biology. EMBO Press, Heidelberg, Germany, Vol. 15, Issue No. 7, Article, 16 pages. https://doi.org/10.15252/msb.20198838
Profile: Prof. Jae Kyoung Kim, PhD
jaekkim@kaist.ac.kr
http://mathsci.kaist.ac.kr/~jaekkim
Associate Professor
Department of Mathematical Sciences
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr Daejeon 34141, Korea
Profile: Dae Wook Kim, PhD Candidate
0308kdo@kaist.ac.kr
http://mathsci.kaist.ac.kr/~jaekkim
PhD Candidate
Department of Mathematical Sciences
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr Daejeon 34141, Korea
Profile: Dr. Cheng Chang, PhD
cheng.chang@pfizer.com
Associate Director of Clinical Pharmacology
Clinical Pharmacology, Global Product Development
Pfizer
https://www.pfizer.com/ Groton 06340, USA
(END)
2019.07.09 View 38228
Mathematical Modeling Makes a Breakthrough for a New CRSD Medication
PhD Candidate Dae Wook Kim (Left) and Professor Jae Kyoung Kim (Right)
- Systems approach reveals photosensitivity and PER2 level as determinants of clock-modulator efficacy -
Mathematicians’ new modeling has identified major sources of interspecies and inter-individual variations in the clinical efficacy of a clock-modulating drug: photosensitivity and PER2 level. This enabled precision medicine for circadian disruption.
A KAIST mathematics research team led by Professor Jae Kyoung Kim, in collaboration with Pfizer, applied a combination of mathematical modeling and simulation tools for circadian rhythms sleep disorders (CRSDs) to analyze the animal data generated by Pfizer. This study was reported in Molecular Systems Biology as the cover article on July 8.
Pharmaceutical companies have conducted extensive studies on animals to determine the candidacy of this new medication. However, the results of animal testing do not always translate to the same effects in human trials. Furthermore, even between humans, efficacy differs across individuals depending on an individual’s genetic and environmental factors, which require different treatment strategies.
To overcome these obstacles, KAIST mathematicians and their collaborators developed adaptive chronotherapeutics to identify precise dosing regimens that could restore normal circadian phase under different conditions.
A circadian rhythm is a 24-hour cycle in the physiological processes of living creatures, including humans. A biological clock in the hypothalamic suprachiasmatic nucleus in the human brain sets the time for various human behaviors such as sleep.
A disruption of the endogenous timekeeping system caused by changes in one’s life pattern leads to advanced or delayed sleep-wake cycle phase and a desynchronization between sleep-wake rhythms, resulting in CRSDs. To restore the normal timing of sleep, timing of the circadian clock could be adjusted pharmacologically.
Pfizer identified PF-670462, which can adjust the timing of circadian clock by inhibiting the core clock kinase of the circadian clock (CK1d/e). However, the efficacy of PF-670462 significantly differs between nocturnal mice and diurnal monkeys, whose sleeping times are opposite.
The research team discovered the source of such interspecies variations in drug response by performing thousands of virtual experiments using a mathematical model, which describes biochemical interactions among clock molecules and PF-670462. The result suggests that the effect of PF-670462 is reduced by light exposure in diurnal primates more than in nocturnal mice. This indicates that the strong counteracting effect of light must be considered in order to effectively regulate the circadian clock of diurnal humans using PF-670462.
Furthermore, the team also found the source of inter-patients variations in drug efficacy using virtual patients whose circadian clocks were disrupted due to various mutations. The degree of perturbation in the endogenous level of the core clock molecule PER2 affects the efficacy.
This explains why the clinical outcomes of clock-modulating drugs are highly variable and certain subtypes are unresponsive to treatment. Furthermore, this points out the limitations of current treatment strategies tailored to only the patient’s sleep and wake time but not to the molecular cause of sleep disorders.
PhD candidate Dae Wook Kim, who is the first author, said that this motivates the team to develop an adaptive chronotherapy, which identifies a personalized optimal dosing time of day by tracking the sleep-wake up time of patients via a wearable device and allows for a precision medicine approach for CRSDs.
Professor Jae Kyoung Kim said, "As a mathematician, I am excited to help enable the advancement of a new drug candidate, which can improve the lives of so many patients. I hope this result promotes more collaborations in this translational research.”
This research was supported by a Pfizer grant to KAIST (G01160179), the Human Frontiers Science Program Organization (RGY0063/2017), and a National Research Foundation (NRF) of Korea Grant (NRF-2016 RICIB 3008468 and NRF-2017-Fostering Core Leaders of the Future Basic Science Program/ Global Ph.D. Fellowship Program).
Figure 1. Interspecies and Inter-patients Variations in PF-670462 Efficacy
Figure 2. Journal Cover Page
Publication:
Dae Wook Kim, Cheng Chang, Xian Chen, Angela C Doran, Francois Gaudreault, Travis Wager, George J DeMarco, and Jae Kyoung Kim. 2019. Systems approach reveals photosensitivity and PER2 level as determinants of clock-modulator efficacy. Molecular Systems Biology. EMBO Press, Heidelberg, Germany, Vol. 15, Issue No. 7, Article, 16 pages. https://doi.org/10.15252/msb.20198838
Profile: Prof. Jae Kyoung Kim, PhD
jaekkim@kaist.ac.kr
http://mathsci.kaist.ac.kr/~jaekkim
Associate Professor
Department of Mathematical Sciences
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr Daejeon 34141, Korea
Profile: Dae Wook Kim, PhD Candidate
0308kdo@kaist.ac.kr
http://mathsci.kaist.ac.kr/~jaekkim
PhD Candidate
Department of Mathematical Sciences
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr Daejeon 34141, Korea
Profile: Dr. Cheng Chang, PhD
cheng.chang@pfizer.com
Associate Director of Clinical Pharmacology
Clinical Pharmacology, Global Product Development
Pfizer
https://www.pfizer.com/ Groton 06340, USA
(END)
2019.07.09 View 38228 -
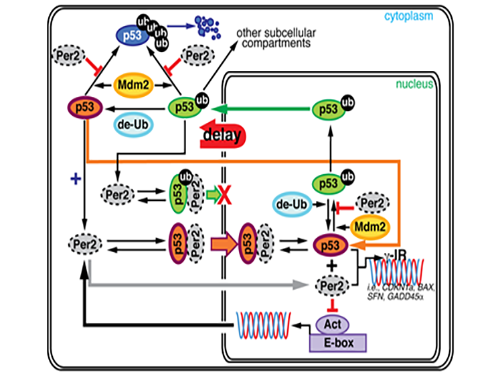 Key Interaction between the Circadian Clock and Cancer Identified
Professor Jae Kyoung Kim and his research team from the Department of Mathematical Sciences at KAIST found that the circadian clock drives changes in circadian rhythms of p53 which functions as a tumor suppressor. Using a differential equation, he applied a model-driven mathematical approach to learn the mechanism and role of p53.
Kim’s mathematical modeling has been validated by experimental studies conducted by a research team at Virginia Polytechnic Institute and State University (Virginia Tech) in the United State, which is led by Professor Carla Finkielstein. As a result, the researchers revealed that there is an important link existed between the circadian clock and cancer.
The findings of this research were published online in Proceedings of the National Academy of Sciences of the United States of the America (PNAS) on November 9, 2016.
The circadian clock in our brain controls behavioral and physiological processes within a period of 24 hours, including making us fall asleep at a certain time by triggering the release of the sleep hormone melatonin in our brain, for example, around 9 pm. The clock is also involved in various physiological processes such as cell division, movement, and development.
Disruptions caused by the mismatch of the circadian clock and real time due to chronic late night work, shiftwork, and other similar issues may lead to various diseases such as diabetes, cancer, and heart disease.
In 2014, when Kim met with Finkielstein, her research team succeeded in observing the changes of p53 over a period of 24 hours, but could not understand how the circadian clock controls the 24-hour rhythm of p53. It was difficult to determine p53’s mechanism since its cell regulation system is far more complex than other cells
To solve the problem, Kim set up a computer simulation using mathematical modeling and ran millions of simulations. Instead of the traditional method based on trial and error experiments, mathematical modeling allowed to save a great deal of time, cost, and manpower.
During this process, Kim proved that the biorhythm of p53 and Period2, an important protein in the circadian clock, are closely related. Cells usually consist of a cell nucleus and cytoplasm. While p53 exists in both nucleus and cytoplasm, it becomes more stable and its degradation slows down when it is in the nucleus.
Kim predicted that the Period2 protein, which plays a key role in the functioning of the circadian clock, could influence the nucleus entry of the p53 protein.
Kim’s predictions based on mathematical modeling have been validated by the Virginia team, thereby revealing a strong connection between the circadian clock and cancer.
Researchers said that this research will help explain the cause of different results from numerous anticancer drugs, which are used to normalize the level of p53, when they are administrated at different times and find the most effective dosing times for the drugs.
They also believe that this study will play an important role in identifying the cause of increasing cancer rates in shift-workers whose circadian clocks are unstable and will contribute to the development of more effective treatments for cancer.
Professor Kim said, “This is an exciting thing that my research can contribute to improving the healthy lives of nurses, police officers, firefighters, and the like, who work in shifts against their circadian rhythms. Taking these findings as an opportunity, I hope to see more active interchanges of ideas between biological sciences and mathematical science in Korea.”
This research has been jointly conducted between KAIST and Virginia Tech and supported by the T. J. Park Science Fellowship of POSCO, the National Science Foundation of the United States, and the Young Researcher Program of the National Research Foundation of Korea.
Picture 1. The complex interaction between tumor antigen p53 and Period2 (Per2) which plays a major role in the circadian clock as revealed by mathematical simulations and experiments
Picture 2. A portion of the mathematical model used in the research
Picture 3. Professor Jae Kyoung Kim (third from left) and the Virginia Tech Research Team
2016.11.17 View 8862
Key Interaction between the Circadian Clock and Cancer Identified
Professor Jae Kyoung Kim and his research team from the Department of Mathematical Sciences at KAIST found that the circadian clock drives changes in circadian rhythms of p53 which functions as a tumor suppressor. Using a differential equation, he applied a model-driven mathematical approach to learn the mechanism and role of p53.
Kim’s mathematical modeling has been validated by experimental studies conducted by a research team at Virginia Polytechnic Institute and State University (Virginia Tech) in the United State, which is led by Professor Carla Finkielstein. As a result, the researchers revealed that there is an important link existed between the circadian clock and cancer.
The findings of this research were published online in Proceedings of the National Academy of Sciences of the United States of the America (PNAS) on November 9, 2016.
The circadian clock in our brain controls behavioral and physiological processes within a period of 24 hours, including making us fall asleep at a certain time by triggering the release of the sleep hormone melatonin in our brain, for example, around 9 pm. The clock is also involved in various physiological processes such as cell division, movement, and development.
Disruptions caused by the mismatch of the circadian clock and real time due to chronic late night work, shiftwork, and other similar issues may lead to various diseases such as diabetes, cancer, and heart disease.
In 2014, when Kim met with Finkielstein, her research team succeeded in observing the changes of p53 over a period of 24 hours, but could not understand how the circadian clock controls the 24-hour rhythm of p53. It was difficult to determine p53’s mechanism since its cell regulation system is far more complex than other cells
To solve the problem, Kim set up a computer simulation using mathematical modeling and ran millions of simulations. Instead of the traditional method based on trial and error experiments, mathematical modeling allowed to save a great deal of time, cost, and manpower.
During this process, Kim proved that the biorhythm of p53 and Period2, an important protein in the circadian clock, are closely related. Cells usually consist of a cell nucleus and cytoplasm. While p53 exists in both nucleus and cytoplasm, it becomes more stable and its degradation slows down when it is in the nucleus.
Kim predicted that the Period2 protein, which plays a key role in the functioning of the circadian clock, could influence the nucleus entry of the p53 protein.
Kim’s predictions based on mathematical modeling have been validated by the Virginia team, thereby revealing a strong connection between the circadian clock and cancer.
Researchers said that this research will help explain the cause of different results from numerous anticancer drugs, which are used to normalize the level of p53, when they are administrated at different times and find the most effective dosing times for the drugs.
They also believe that this study will play an important role in identifying the cause of increasing cancer rates in shift-workers whose circadian clocks are unstable and will contribute to the development of more effective treatments for cancer.
Professor Kim said, “This is an exciting thing that my research can contribute to improving the healthy lives of nurses, police officers, firefighters, and the like, who work in shifts against their circadian rhythms. Taking these findings as an opportunity, I hope to see more active interchanges of ideas between biological sciences and mathematical science in Korea.”
This research has been jointly conducted between KAIST and Virginia Tech and supported by the T. J. Park Science Fellowship of POSCO, the National Science Foundation of the United States, and the Young Researcher Program of the National Research Foundation of Korea.
Picture 1. The complex interaction between tumor antigen p53 and Period2 (Per2) which plays a major role in the circadian clock as revealed by mathematical simulations and experiments
Picture 2. A portion of the mathematical model used in the research
Picture 3. Professor Jae Kyoung Kim (third from left) and the Virginia Tech Research Team
2016.11.17 View 8862 -
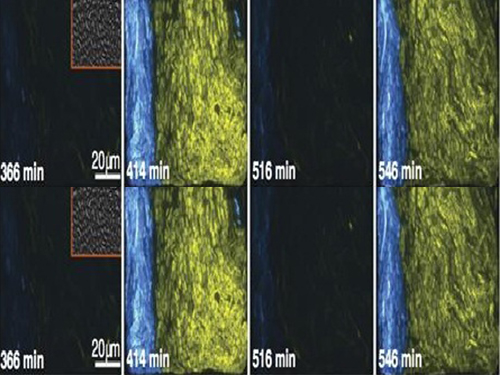 KAIST's Mathematician Reveals the Mechanism for Sustaining Biological Rhythms
Our bodies have a variety of biological clocks that follow rhythms or oscillations with periods ranging from seconds to days. For example, our hearts beat every second, and cells divide periodically. The circadian clock located in the hypothalamus generates twenty-four hour rhythms, timing our sleep and hormone release. How do these biological clocks or circuits generate and sustain the stable rhythms that are essential to life?
Jae Kyoung Kim, who is an assistant professor in the Department of Mathematical Sciences at KAIST, has predicted how these biological circuits generate rhythms and control their robustness, utilizing mathematical modeling based on differential equations and stochastic parameter sampling. Based on his prediction, using synthetic biology, a research team headed by Matthew Bennett of Rice University constructed a novel biological circuit that spans two genetically engineered strains of bacteria, one serves as an activator and the other as a repressor to regulate gene expression across multiple cell types, and found that the circuit generates surprisingly robust rhythms under various conditions.
The results of the research conducted in collaboration with KAIST (Korea Institute of Science and Technology), Rice University, and the University of Houston were published in Science (August 28, 2015 issue). The title of the paper is "Emergent Genetic Oscillations in a Synthetic Microbial Consortium" .
The top-down research approach, which focuses on identifying the components of biological circuits, limits our understanding of the mechanisms in which the circuits generate rhythms. Synthetic biology, a rapidly growing field at the interface of biosciences and engineering, however, uses a bottom-up approach.
Synthetic biologists can create complex circuits out of simpler components, and some of these new genetic circuits are capable of fluctuation to regulate gene production. In the same way that electrical engineers understand how an electrical circuit works as they construct batteries, resistors, and wires, synthetic biologists can understand better about biological circuits if they put them together using genes and proteins. However, due to the complexity of biological systems, both experiments and mathematical modeling need to be applied hand in hand to design these biological circuits and understand their function.
In this research, an interdisciplinary approach proved that a synthetic intercellular singling circuit generates robust rhythms to create a cooperative microbial system. Specifically, Kim's mathematical analysis suggested, and experiments confirmed, that the presence of negative feedback loops in addition to a core transcriptional negative feedback loop can explain the robustness of rhythms in this system. This result provides important clues about the fundamental mechanism of robust rhythm generation in biological systems.
Furthermore, rather than constructing the entire circuit inside a single bacterial strain, the circuit was split among two strains of Escherichia coli bacterium. When the strains were grown together, the bacteria exchanged information, completing the circuit. Thus, this research also shows how, by regulating individual cells within the system, complex biological systems can be controlled, which in turn influences each other (e.g., the gut microbiome in humans).
###
Ye Chen, a graduate student in Bennett's laboratory at Rice University, and Jae Kyoung Kim, an assistant professor at KAIST and a former postdoctoral fellow at Ohio State University, are the lead authors of the paper. The co-authors are Rice graduate student Andrew Hirning and Krešimir Josic?, a professor of mathematics at the University of Houston. Bennett is the Assistant Professor of the Biochemistry and Cell Biology Department at Rice University.
About the researcher: While Jae Kyoung Kim is a mathematician, he has also solved various biological puzzles in collaboration with various experimental laboratories of Matthew Bennett at Rice University, David Virshup at Duke and the National University of Singapore, Carla Finkielstein at Virginia Polytechnic Institute and State University, Choo-Gon Lee at the Florida State University, Seung-Hee Yoo at the Medical School of the University of Texas, Toru Takumi at RIKEN Brain Science Institute, and Travis Wager at Pfizer Inc. He has used non-linear dynamics and stochastic analysis to understand the function of biochemical networks in biological systems. In particular, he is interested in mechanisms generating and regulating biological rhythms.
Picture 1: This schematic image is the design of a biological circuit between two strains of bacteria and the part of differential equations used to understand the function of the biological circuit.
Picture 2: The core transcriptional negative feedback loop and additional negative feedback loop in the biological circuit (picture 1) generate robust rhythms. The snapshots correspond the red dots in the time series graph.
2015.08.31 View 9715
KAIST's Mathematician Reveals the Mechanism for Sustaining Biological Rhythms
Our bodies have a variety of biological clocks that follow rhythms or oscillations with periods ranging from seconds to days. For example, our hearts beat every second, and cells divide periodically. The circadian clock located in the hypothalamus generates twenty-four hour rhythms, timing our sleep and hormone release. How do these biological clocks or circuits generate and sustain the stable rhythms that are essential to life?
Jae Kyoung Kim, who is an assistant professor in the Department of Mathematical Sciences at KAIST, has predicted how these biological circuits generate rhythms and control their robustness, utilizing mathematical modeling based on differential equations and stochastic parameter sampling. Based on his prediction, using synthetic biology, a research team headed by Matthew Bennett of Rice University constructed a novel biological circuit that spans two genetically engineered strains of bacteria, one serves as an activator and the other as a repressor to regulate gene expression across multiple cell types, and found that the circuit generates surprisingly robust rhythms under various conditions.
The results of the research conducted in collaboration with KAIST (Korea Institute of Science and Technology), Rice University, and the University of Houston were published in Science (August 28, 2015 issue). The title of the paper is "Emergent Genetic Oscillations in a Synthetic Microbial Consortium" .
The top-down research approach, which focuses on identifying the components of biological circuits, limits our understanding of the mechanisms in which the circuits generate rhythms. Synthetic biology, a rapidly growing field at the interface of biosciences and engineering, however, uses a bottom-up approach.
Synthetic biologists can create complex circuits out of simpler components, and some of these new genetic circuits are capable of fluctuation to regulate gene production. In the same way that electrical engineers understand how an electrical circuit works as they construct batteries, resistors, and wires, synthetic biologists can understand better about biological circuits if they put them together using genes and proteins. However, due to the complexity of biological systems, both experiments and mathematical modeling need to be applied hand in hand to design these biological circuits and understand their function.
In this research, an interdisciplinary approach proved that a synthetic intercellular singling circuit generates robust rhythms to create a cooperative microbial system. Specifically, Kim's mathematical analysis suggested, and experiments confirmed, that the presence of negative feedback loops in addition to a core transcriptional negative feedback loop can explain the robustness of rhythms in this system. This result provides important clues about the fundamental mechanism of robust rhythm generation in biological systems.
Furthermore, rather than constructing the entire circuit inside a single bacterial strain, the circuit was split among two strains of Escherichia coli bacterium. When the strains were grown together, the bacteria exchanged information, completing the circuit. Thus, this research also shows how, by regulating individual cells within the system, complex biological systems can be controlled, which in turn influences each other (e.g., the gut microbiome in humans).
###
Ye Chen, a graduate student in Bennett's laboratory at Rice University, and Jae Kyoung Kim, an assistant professor at KAIST and a former postdoctoral fellow at Ohio State University, are the lead authors of the paper. The co-authors are Rice graduate student Andrew Hirning and Krešimir Josic?, a professor of mathematics at the University of Houston. Bennett is the Assistant Professor of the Biochemistry and Cell Biology Department at Rice University.
About the researcher: While Jae Kyoung Kim is a mathematician, he has also solved various biological puzzles in collaboration with various experimental laboratories of Matthew Bennett at Rice University, David Virshup at Duke and the National University of Singapore, Carla Finkielstein at Virginia Polytechnic Institute and State University, Choo-Gon Lee at the Florida State University, Seung-Hee Yoo at the Medical School of the University of Texas, Toru Takumi at RIKEN Brain Science Institute, and Travis Wager at Pfizer Inc. He has used non-linear dynamics and stochastic analysis to understand the function of biochemical networks in biological systems. In particular, he is interested in mechanisms generating and regulating biological rhythms.
Picture 1: This schematic image is the design of a biological circuit between two strains of bacteria and the part of differential equations used to understand the function of the biological circuit.
Picture 2: The core transcriptional negative feedback loop and additional negative feedback loop in the biological circuit (picture 1) generate robust rhythms. The snapshots correspond the red dots in the time series graph.
2015.08.31 View 9715