drug+interactions
-
 KAIST leads AI-based analysis on drug-drug interactions involving Paxlovid
KAIST (President Kwang Hyung Lee) announced on the 16th that an advanced AI-based drug interaction prediction technology developed by the Distinguished Professor Sang Yup Lee's research team in the Department of Biochemical Engineering that analyzed the interaction between the PaxlovidTM ingredients that are used as COVID-19 treatment and other prescription drugs was published as a thesis. This paper was published in the online edition of 「Proceedings of the National Academy of Sciences of America」 (PNAS), an internationally renowned academic journal, on the 13th of March.
* Thesis Title: Computational prediction of interactions between Paxlovid and prescription drugs (Authored by Yeji Kim (KAIST, co-first author), Jae Yong Ryu (Duksung Women's University, co-first author), Hyun Uk Kim (KAIST, co-first author), and Sang Yup Lee (KAIST, corresponding author))
In this study, the research team developed DeepDDI2, an advanced version of DeepDDI, an AI-based drug interaction prediction model they developed in 2018. DeepDDI2 is able to compute for and process a total of 113 drug-drug interaction (DDI) types, more than the 86 DDI types covered by the existing DeepDDI.
The research team used DeepDDI2 to predict possible interactions between the ingredients (ritonavir, nirmatrelvir) of Paxlovid*, a COVID-19 treatment, and other prescription drugs. The research team said that while among COVID-19 patients, high-risk patients with chronic diseases such as high blood pressure and diabetes are likely to be taking other drugs, drug-drug interactions and adverse drug reactions for Paxlovid have not been sufficiently analyzed, yet. This study was pursued in light of seeing how continued usage of the drug may lead to serious and unwanted complications.
* Paxlovid: Paxlovid is a COVID-19 treatment developed by Pfizer, an American pharmaceutical company, and received emergency use approval (EUA) from the US Food and Drug Administration (FDA) in December 2021.
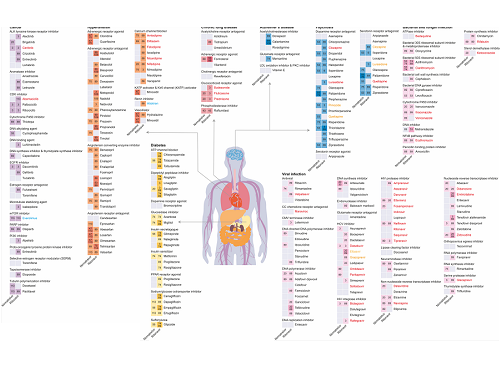
The research team used DeepDDI2 to predict how Paxrovid's components, ritonavir and nirmatrelvir, would interact with 2,248 prescription drugs. As a result of the prediction, ritonavir was predicted to interact with 1,403 prescription drugs and nirmatrelvir with 673 drugs.
Using the prediction results, the research team proposed alternative drugs with the same mechanism but low drug interaction potential for prescription drugs with high adverse drug events (ADEs). Accordingly, 124 alternative drugs that could reduce the possible adverse DDI with ritonavir and 239 alternative drugs for nirmatrelvir were identified.
Through this research achievement, it became possible to use an deep learning technology to accurately predict drug-drug interactions (DDIs), and this is expected to play an important role in the digital healthcare, precision medicine and pharmaceutical industries by providing useful information in the process of developing new drugs and making prescriptions.
Distinguished Professor Sang Yup Lee said, "The results of this study are meaningful at times like when we would have to resort to using drugs that are developed in a hurry in the face of an urgent situations like the COVID-19 pandemic, that it is now possible to identify and take necessary actions against adverse drug reactions caused by drug-drug interactions very quickly.”
This research was carried out with the support of the KAIST New-Deal Project for COVID-19 Science and Technology and the Bio·Medical Technology Development Project supported by the Ministry of Science and ICT.
Figure 1. Results of drug interaction prediction between Paxlovid ingredients and representative approved drugs using DeepDDI2
2023.03.16 View 8688
KAIST leads AI-based analysis on drug-drug interactions involving Paxlovid
KAIST (President Kwang Hyung Lee) announced on the 16th that an advanced AI-based drug interaction prediction technology developed by the Distinguished Professor Sang Yup Lee's research team in the Department of Biochemical Engineering that analyzed the interaction between the PaxlovidTM ingredients that are used as COVID-19 treatment and other prescription drugs was published as a thesis. This paper was published in the online edition of 「Proceedings of the National Academy of Sciences of America」 (PNAS), an internationally renowned academic journal, on the 13th of March.
* Thesis Title: Computational prediction of interactions between Paxlovid and prescription drugs (Authored by Yeji Kim (KAIST, co-first author), Jae Yong Ryu (Duksung Women's University, co-first author), Hyun Uk Kim (KAIST, co-first author), and Sang Yup Lee (KAIST, corresponding author))
In this study, the research team developed DeepDDI2, an advanced version of DeepDDI, an AI-based drug interaction prediction model they developed in 2018. DeepDDI2 is able to compute for and process a total of 113 drug-drug interaction (DDI) types, more than the 86 DDI types covered by the existing DeepDDI.
The research team used DeepDDI2 to predict possible interactions between the ingredients (ritonavir, nirmatrelvir) of Paxlovid*, a COVID-19 treatment, and other prescription drugs. The research team said that while among COVID-19 patients, high-risk patients with chronic diseases such as high blood pressure and diabetes are likely to be taking other drugs, drug-drug interactions and adverse drug reactions for Paxlovid have not been sufficiently analyzed, yet. This study was pursued in light of seeing how continued usage of the drug may lead to serious and unwanted complications.
* Paxlovid: Paxlovid is a COVID-19 treatment developed by Pfizer, an American pharmaceutical company, and received emergency use approval (EUA) from the US Food and Drug Administration (FDA) in December 2021.
The research team used DeepDDI2 to predict how Paxrovid's components, ritonavir and nirmatrelvir, would interact with 2,248 prescription drugs. As a result of the prediction, ritonavir was predicted to interact with 1,403 prescription drugs and nirmatrelvir with 673 drugs.
Using the prediction results, the research team proposed alternative drugs with the same mechanism but low drug interaction potential for prescription drugs with high adverse drug events (ADEs). Accordingly, 124 alternative drugs that could reduce the possible adverse DDI with ritonavir and 239 alternative drugs for nirmatrelvir were identified.
Through this research achievement, it became possible to use an deep learning technology to accurately predict drug-drug interactions (DDIs), and this is expected to play an important role in the digital healthcare, precision medicine and pharmaceutical industries by providing useful information in the process of developing new drugs and making prescriptions.
Distinguished Professor Sang Yup Lee said, "The results of this study are meaningful at times like when we would have to resort to using drugs that are developed in a hurry in the face of an urgent situations like the COVID-19 pandemic, that it is now possible to identify and take necessary actions against adverse drug reactions caused by drug-drug interactions very quickly.”
This research was carried out with the support of the KAIST New-Deal Project for COVID-19 Science and Technology and the Bio·Medical Technology Development Project supported by the Ministry of Science and ICT.
Figure 1. Results of drug interaction prediction between Paxlovid ingredients and representative approved drugs using DeepDDI2
2023.03.16 View 8688 -
 Deep Learning Predicts Drug-Drug and Drug-Food Interactions
A Korean research team from KAIST developed a computational framework, DeepDDI, that accurately predicts and generates 86 types of drug-drug and drug-food interactions as outputs of human-readable sentences, which allows in-depth understanding of the drug-drug and drug-food interactions.
Drug interactions, including drug-drug interactions (DDIs) and drug-food constituent interactions (DFIs), can trigger unexpected pharmacological effects, including adverse drug events (ADEs), with causal mechanisms often unknown. However, current prediction methods do not provide sufficient details beyond the chance of DDI occurrence, or require detailed drug information often unavailable for DDI prediction.
To tackle this problem, Dr. Jae Yong Ryu, Assistant Professor Hyun Uk Kim and Distinguished Professor Sang Yup Lee, all from the Department of Chemical and Biomolecular Engineering at Korea Advanced Institute of Science and Technology (KAIST), developed a computational framework, named DeepDDI, that accurately predicts 86 DDI types for a given drug pair. The research results were published online in Proceedings of the National Academy of Sciences of the United States of America (PNAS) on April 16, 2018, which is entitled “Deep learning improves prediction of drug-drug and drug-food interactions.”
DeepDDI takes structural information and names of two drugs in pair as inputs, and predicts relevant DDI types for the input drug pair. DeepDDI uses deep neural network to predict 86 DDI types with a mean accuracy of 92.4% using the DrugBank gold standard DDI dataset covering 192,284 DDIs contributed by 191,878 drug pairs. Very importantly, DDI types predicted by DeepDDI are generated in the form of human-readable sentences as outputs, which describe changes in pharmacological effects and/or the risk of ADEs as a result of the interaction between two drugs in pair. For example, DeepDDI output sentences describing potential interactions between oxycodone (opioid pain medication) and atazanavir (antiretroviral medication) were generated as follows: “The metabolism of Oxycodone can be decreased when combined with Atazanavir”; and “The risk or severity of adverse effects can be increased when Oxycodone is combined with Atazanavir”. By doing this, DeepDDI can provide more specific information on drug interactions beyond the occurrence chance of DDIs or ADEs typically reported to date.
DeepDDI was first used to predict DDI types of 2,329,561 drug pairs from all possible combinations of 2,159 approved drugs, from which DDI types of 487,632 drug pairs were newly predicted. Also, DeepDDI can be used to suggest which drug or food to avoid during medication in order to minimize the chance of adverse drug events or optimize the drug efficacy. To this end, DeepDDI was used to suggest potential causal mechanisms for the reported ADEs of 9,284 drug pairs, and also predict alternative drug candidates for 62,707 drug pairs having negative health effects to keep only the beneficial effects. Furthermore, DeepDDI was applied to 3,288,157 drug-food constituent pairs (2,159 approved drugs and 1,523 well-characterized food constituents) to predict DFIs. The effects of 256 food constituents on pharmacological effects of interacting drugs and bioactivities of 149 food constituents were also finally predicted. All these prediction results can be useful if an individual is taking medications for a specific (chronic) disease such as hypertension or diabetes mellitus type 2.
Distinguished Professor Sang Yup Lee said, “We have developed a platform technology DeepDDI that will allow precision medicine in the era of Fourth Industrial Revolution. DeepDDI can serve to provide important information on drug prescription and dietary suggestions while taking certain drugs to maximize health benefits and ultimately help maintain a healthy life in this aging society.”
Figure 1. Overall scheme of Deep DDDI and prediction of food constituents that reduce the in vivo concentration of approved drugs
2018.04.18 View 12616
Deep Learning Predicts Drug-Drug and Drug-Food Interactions
A Korean research team from KAIST developed a computational framework, DeepDDI, that accurately predicts and generates 86 types of drug-drug and drug-food interactions as outputs of human-readable sentences, which allows in-depth understanding of the drug-drug and drug-food interactions.
Drug interactions, including drug-drug interactions (DDIs) and drug-food constituent interactions (DFIs), can trigger unexpected pharmacological effects, including adverse drug events (ADEs), with causal mechanisms often unknown. However, current prediction methods do not provide sufficient details beyond the chance of DDI occurrence, or require detailed drug information often unavailable for DDI prediction.
To tackle this problem, Dr. Jae Yong Ryu, Assistant Professor Hyun Uk Kim and Distinguished Professor Sang Yup Lee, all from the Department of Chemical and Biomolecular Engineering at Korea Advanced Institute of Science and Technology (KAIST), developed a computational framework, named DeepDDI, that accurately predicts 86 DDI types for a given drug pair. The research results were published online in Proceedings of the National Academy of Sciences of the United States of America (PNAS) on April 16, 2018, which is entitled “Deep learning improves prediction of drug-drug and drug-food interactions.”
DeepDDI takes structural information and names of two drugs in pair as inputs, and predicts relevant DDI types for the input drug pair. DeepDDI uses deep neural network to predict 86 DDI types with a mean accuracy of 92.4% using the DrugBank gold standard DDI dataset covering 192,284 DDIs contributed by 191,878 drug pairs. Very importantly, DDI types predicted by DeepDDI are generated in the form of human-readable sentences as outputs, which describe changes in pharmacological effects and/or the risk of ADEs as a result of the interaction between two drugs in pair. For example, DeepDDI output sentences describing potential interactions between oxycodone (opioid pain medication) and atazanavir (antiretroviral medication) were generated as follows: “The metabolism of Oxycodone can be decreased when combined with Atazanavir”; and “The risk or severity of adverse effects can be increased when Oxycodone is combined with Atazanavir”. By doing this, DeepDDI can provide more specific information on drug interactions beyond the occurrence chance of DDIs or ADEs typically reported to date.
DeepDDI was first used to predict DDI types of 2,329,561 drug pairs from all possible combinations of 2,159 approved drugs, from which DDI types of 487,632 drug pairs were newly predicted. Also, DeepDDI can be used to suggest which drug or food to avoid during medication in order to minimize the chance of adverse drug events or optimize the drug efficacy. To this end, DeepDDI was used to suggest potential causal mechanisms for the reported ADEs of 9,284 drug pairs, and also predict alternative drug candidates for 62,707 drug pairs having negative health effects to keep only the beneficial effects. Furthermore, DeepDDI was applied to 3,288,157 drug-food constituent pairs (2,159 approved drugs and 1,523 well-characterized food constituents) to predict DFIs. The effects of 256 food constituents on pharmacological effects of interacting drugs and bioactivities of 149 food constituents were also finally predicted. All these prediction results can be useful if an individual is taking medications for a specific (chronic) disease such as hypertension or diabetes mellitus type 2.
Distinguished Professor Sang Yup Lee said, “We have developed a platform technology DeepDDI that will allow precision medicine in the era of Fourth Industrial Revolution. DeepDDI can serve to provide important information on drug prescription and dietary suggestions while taking certain drugs to maximize health benefits and ultimately help maintain a healthy life in this aging society.”
Figure 1. Overall scheme of Deep DDDI and prediction of food constituents that reduce the in vivo concentration of approved drugs
2018.04.18 View 12616